Research Article, Vegetos Vol: 29 Issue: 1
Marker based Genetic Diversity of Rice Genotypes for Salinity Tolerance at Panicle Initiation Stage
| Sala M1*, Shanthi P1, Selvi B2 and Ravi V1 | |
| 1Tamil Nadu Rice Research Institute, Aduthurai, Tamilnadu Agriculture University (TNAU), Tamilnadu, India | |
| 2Department of Millets, Centre for Plant Breeding & Genetics, TNAU, Coimbatore, Tamilnadu, India | |
| Corresponding author : Sala M Tamil Nadu Rice Research Institute, Aduthurai, TNAU, Tamilnadu, India Tel: 8012460114 E-mail: swtsala1@gmail.com |
|
| Received: December 22, 2015 Accepted: February 04, 2016 Published: February 11, 2016 | |
| Citation: Sala M, Shanthi P, Selvi B, Ravi V (2016) Marker based Genetic Diversity of Rice Genotypes for Salinity Tolerance at Panicle Initiation Stage. Vegetos 29:1. doi: 10.5958/2229-4473.2016.00004.5 |
Abstract
Marker based Genetic Diversity of Rice Genotypes for Salinity Tolerance at Panicle Initiation Stage
The study was designed to evaluate salinity tolerance at panicle initiation stage and characterize the genetic diversity in a set of rice genotypes with different adaptation to saline soil using microsatellite markers (SSR markers). In evaluation for salinity tolerance the land races Nona bokra and Pokkali showed high tolerance at 6 EC dsm- 1 (Electrical Conductivity) level. In panicle initiation stage followed by FL 478 and CSR 36 with moderate tolerance. For diversity analysis, a total of 70 SSR primers across the 12 chromosome were used. Of these, 34 were polymorphic among the selected genotypes. The diversity analysis grouped the 13 genotypes into six clusters. Cluster I consisted of three varieties viz., ADT37, ADT 47 and TRY(R)3. Cluster II was the biggest cluster having six varieties viz., ADT42, ADT43, AD09225, FL478, CSR10 and CSR36. Cluster III consisted of mono cluster of TNAU RiceADT49 and Cluster IV consisted of TRY(R)2. Cluster V consisted of monocluster of Pokkali and cluster VI consist of Nonabokra which is highly salt tolerant. The maximum similarity value was 0.828 and the minimum similarity value was 0.502. The genetic distance was ranged between 0.172 to 0.489. The lower genetic distance of 0.172 was observed between FL478 and AD09225, and the higher genetic distance was observed between Nona bokra and ADT47 (0.489) followed by TRY(R)2 and Pokkali. The Polymorphism information content ranged from 0.138 to 0.705. The highest PIC value observed in RM493 (0.705) followed by RM 3412 (0.638) and RM 412(0.591). The lowest PIC value was observed in RM5933 (0.118), RM 240 (0.142) and RM 287 (0.138). Hence parental selection based on the genetic diversity is highly essential to develop a good variety. In future by utilizing the highly genetic divergent parents of ADT(R)47 and Nonabokra may help to develop high yielding salt tolerant varieties.
Keywords: Rice; Microsatellite markers; Cluster; Similarity value; Genetic distance
Keywords |
|
| Rice; Microsatellite markers; Cluster; Similarity value; Genetic distance | |
Introduction |
|
| Rice is the staple food for one third of the world’s population and occupies almost one-fifth of the total land area covered under cereals [1]. By the year 2025, 21 % increase in rice production will be needed over that of year 2000 to meet the food demand of increased population [2]. It is imperative to increase rice production in different rice growing ecosystems including salt affected coastal areas to feed the increasing world population [3]. The area affected by different level of salinity covers about 400-950 million hectares of land around the world [4]. In India a potential area of 20 milllion hectares of land has been affected by varying degrees of salinity. In Tamilnadu one lakhs and thirty thousand hectare are affected by coastal salinity [5]. | |
| Many agronomic practices followed for reclamation practices, cultivation of salt tolerant variety is one of the most economical and possible ways to make use of saline areas. It is quite well known that rice show variation in salt tolerance [6]. Though rice is considered sensitive to salinity, it is one of the few crops that can thrive on saltaffected soils. Rice is relatively tolerant to salinity at the germination stage but it is highly sensitive at seedling and panicle initiation stage. Panicle initiation stage is directly related to crop yield [7]. Stress at panicle initiation stage causes spikelet sterility that ultimately leads to yield loss. | |
| Screening of rice genotypes at seedling stage is comparatively easier and also more rapid than reproductive stage [8]. A number of morpho-physiological growth factors are affected by NaCl stress [9]. Hence breeding for salt tolerance in rice is difficult and very slow due to complexity of this polygenic trait and insufficient knowledge about mechanism of inheritance of genes controlling the character, low genetic variance at different developmental and physiological stages. | |
| In recent years, a rapid advancement has been made towards the development of molecular marker technology and their application in plant breeding programs [10]. SSR or microsatellite markers are proved to be ideal for making genetic maps [11,12], assisting selection [13] and studying genetic diversity in different genotypes. During the last two decades, these markers helped in mapping several rice genes/QTLs for salt tolerant parameters like Na+ and K+ uptake, Na+ and K+ concentration and Na+/K+ ratio in shoot. Based on these findings, Saltol (for seedling tolerance) is a major QTL mapped on the short arm of chromosome [8]. Later studies confirmed that Saltol controlled Na-K absorption. During the reproductive period of rice, salinity causes morphological changes similar to other environmental stresses that cause growth inhibition of plant structures, such as degeneration of primary and second panicle. This stage directly influence the yield of the crop. Hence development of tolerance for salinity at reproductive stage is highly essential. Based on the above circumstance, the present study aimed to screen rice genotypes under saline conditions for panicle initiation stage and assess its diversity to identify the diverse parents to utilize in breeding programs. | |
Materials and Methods |
|
| Plant materials | |
| A total of 13 genotypes including two salt tolerant land races (Pokkali, Nonabokra), five salt tolerant varieties(TRY2,TRY3,FL4 78,CSR10,CSR36) and six popular high yielding varieties (ADT37, ADT42, ADT43, ADT(R)47, TNAU RiceADT49,ADO9225) were selected for this study. The details of the plant material are described in Table 1. | |
| Table 1: Origin, characteristics and salt tolerance reaction at panicle initiation stage of 13 rice genotypes. | |
| Screening of rice genotypes for salinity tolerance at the reproductive stage | |
| The genotypes were evaluated for their salinity tolerance at panicle initiation stage under artificial screening condition. For this screening, the genotypes at tillering stage were transferred to pots in completely randomized design with three replications. For establishment the nutrient solutions (YOSHIDA) were added in weekly interval [14]. After establishment the seedlings were treated with saline solutions at 6 Electrical Conductivity (ECdsm-1) in weekly interval along with the nutrient solution. For comparison one set of control is maintained under normal condition. The genotypes were observed closely and the observations were tabulated in Table 2. | |
| Table 2: Modified standard evaluating score (SES) salt injury at seedling stage. | |
| Molecular marker analysis | |
| All the selected genotypes were screened with 70 SSR primers [15], including the primers linked to salinity tolerance, in order to conduct diversity analysis. The leaf samples were collected from ten days old seedlings and used for DNA Extraction. The genomic DNA was extracted according to dellaporta with some modifications [16]. The concentration of DNA was measured by running the genomic DNA in 0.8 % agarose gel. Based on the concentrations, the DNA was diluted to the concentration of 50 ng.mL-1 and the PCR reactions were carried out in thermo cycler. The PCR reactions were maintained as described by Panaud et al. [17]. The amplified products were separated in 1.5% agarose gel then stained ethidium bromide (0.8 μg/ mL). The gel was visualized in UV transilluminator and photographs were taken using gel documentation system Syngene bio imaging. | |
| Statistical analysis of SSR data | |
| Each SSR band was scored as present (1), absent (0) or (9) as a missing observation for each genotype. An accession was assigned a null allele for a microsatellite locus whenever an amplification product could not be detected for a particular genotype-marker combination [18]. To measure the informativeness of the markers, the Polymorphic Information Content (PIC) for each SSR locus was calculated according to the formula | |
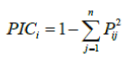 |
|
| Where Pij, is the frequency of the jth allele for the ith marker and summed over n alleles in the set of 13 genotypes investigated. The frequencies of null allele were not included in the calculation of PIC values. Genetic similarity (GS) between genotype I and j was estimated by using Jaccard’s coefficient, as described by Sneath and Sokal [19]. Markers with missing observations for genotype i and j were not included in the calculation of GSij. Based on the genetic similarity matrix, an unweighted paired group method of arithmetic averages (UPGMA) cluster analysis was used to access the pattern of genetic diversity among the rice genotypes. All calculations were performed by using NTSYS pc version 2.1 software [20]. | |
Results and Discussion |
|
| Rice is relatively tolerant to salt stress during germination and maturity stage. However, during the maturity stage (early seedling stage and panicle initiation, anthesis and fertilization), it becomes very sensitive [21]. Studies have shown that a very poor correlation exists between tolerances at seedling stage and reproductive stage, suggesting that tolerance at these two stages is regulated by a different set of genes [22]. | |
| Screening of rice genotypes at panicle initiation stage | |
| All the thirteen tested genotypes had recorded wide variation in tolerance for saline toxicity at 6 EC dsm-1 and its score ranging from 1 (highly tolerant) to 9 (highly susceptible), as shown in Table 2. This scoring was done based on IRRI system of classification as described in materials and methods and Table 2. The two land races viz., Nonabokra and Pokkali, were highly tolerant to salinity, whereas the five salt tolerant varieties viz., FL 478, CSR 36, CSR 10, TRY(R)2 and TRY(R)3 were showed moderately tolerant, and the remaining high yielding popular varieties viz., ADT37, ADT43, ADT(R)47, ADT49, ADT 42 and AD 09225were susceptible. These results confirmed the findings of Mehede Hassan Rubel et al. [23], that FL478 showed moderately tolerance to salinity. Gopikannan [24] identified that ASD 18, IR 20, ADT 49, CO (R) 49 and CO (R) 50 were susceptible to salinity at seedling stage. | |
| Genotypic grouping of rice genotypes using SSR markers | |
| The genetic relationship among the rice genotypes was assessed by UPGMA cluster analysis of similarity matrix representing the Jaccard’s coefficient. Cluster analysis was used to group the genotypes and to construct a dendrogram. The dendrogram revealed that the thirteen genotypes were grouped in to six clusters (Figure 1). The cluster II is the biggest cluster consisting of six genotypes followed by cluster I with three genotypes and Cluster II with six genotypes and cluster III, IV, V and VI are monoclusters. Cluster I consisted of three varieties viz., ADT37, ADT (R)47, TRY(R)3. ADT (R)47 and TRY (R) 3 have the same parentage of ADT 43 and Jeeraga samba. Hence these genotypes are grouped in to same cluster. Cluster II was the biggest cluster consisted of six varieties viz., ADT42, ADT 43, AD09225, FL478, CSR10 and CSR36. Even though ADT 43 is one of the parent of TRY(R)3 and ADT (R) 47, it is grouped into different clusters. This may be due to the recombination and selection intensity of the varieties during hybridization programme. The salt tolerant and susceptible genotypes are grouped in to same cluster indicates they posses more similar characters except the tolerance. Cluster III IV, V and VI are monoclusters consisted of ADT49, TRY (R)2, Pokkali and Nona bokra. This indicates that these genotypes are highly diverse than the other genotypes. Hence crossing these genotypes with other genotypes may give good segregants. Shanthi et al., [25] reported that the salt tolerant landrace Pokkali and CSR 23were grouped into separate cluster. | |
| Figure 1: Genetic similarity analysis using SSR marker in rice genotypes. | |
| Based on the cluster analysis, the similarity values are tabulated in Table 4. The similarity values were ranged from 0.502 to 0.828. The lowest similarity value of 0.506 and the high genetic distance value of 0.489 was observed between ADT (R) 47 and Nona bokra (Table 4). The highest similarity value of 0.828 the low genetic distance of 0.172 was observed between AD09225 and FL 478. High genetic similarity among the genotypes of diverse region were also reported by Chakravarthi and Rambabu [26], Ram et al. [18] and Senguttuvel [27] in their studies using SSR markers. The similarity value in most of the genotypes are more than 50%, indicating medium level of polymorphism observed in our study, compared to 80% similarity of indica genotypes by Chakravathi and Rambabu [26]. Similarly, less similarity value was also reported by Bhuyian et al. [28] among the rice genotypes. | |
| Table 4: Similarity coefficient value and genetic distance calculated for the 13 genotypes based on SSR marker data. | |
| Molecular diversity and salt tolerance | |
| SSR markers are widely used for fingerprinting and diversity studies in rice genotypes due to its high polymorphic rate which can be identified even at individual level. Primers were selected based on their capability to deliver a clear, positive, reproducible, and polymorphic banding pattern in all the genotypes. The 70 primers were used for the diversity analysis. Of these 34 were polymorphic (Table 3). The levels of polymorphism among 13 rice genotypes were evaluated by calculating the PIC value. A total of 89 alleles were detected in 34 polymorphic SSR primers. The maximum of four alleles were recorded in the following primers viz., RM 493,RM3412, RM148 and RM 206; with the three alleles of markers viz.,RM412, RM336, RM 1233, RM210, RM224. The lowest number of alleles (2) was detected in RM5511, RM 220, RM 208. The average number of alleles per loci is 2.61 and total number of alleles are eighty nine (Table 5). Hussian et al. [29] detected a total of 38 alleles in 12 aromatic rice landraces of Bangladesh, and the average number of alleles was 3.8 per locus. | |
| Table 3: List of Polymorphic primers used for this study. | |
| Table 5: Number of alleles and Polymorphic Information Content (PIC) values of SSR markers for 13 rice genotypes. | |
| PIC value is a reflection of allelic diversity and frequency among the varieties. The PIC value ranged from 0.138 to 0.705. The highest PIC value was observed in RM493 (0.705), followed by RM 3412 (0.638) and RM412 (0.591). The lowest PIC value was observed in RM5933 (0.118), RM 240 (0.142) and RM 287 (0.138), and the average PIC value was 0.119. According to Borbora et al. [30], the PIC values varied greatly among markers from 0.19 to 0.90, with an average of 0.75, which is markedly higher than the molecular diversity analysis of stress tolerant rice using SSR marker which was conducted by Islam et al. [31]. Deepti davla et al. [32] reported that the PIC values for 26 SSR markers varied from 0.50 (RM6737) to 0.89(RM3412), with an average PIC of 6.70. | |
| In our results we showed that the key markers viz., RM412, RM 493 RM 3412, RM336, RM17, RM10825, and RM206 are the most informative, as shown by their higher PIC values, as well as their capability to produce greater numbers of polymorphic bands (Figure 2). Similarly, Krishnendu Chattopadhyay et al. [33] reported that primers viz., RM10682, RM10719, RM10745, and RM3412 proved to be more informative, as shown by their high PIC and MI values, as well as their capability to produce greater numbers of polymorphic bands [34]. Mehede Hassan Rubel et al. [23] found that three primers (RM10772, RM7075 and RM296) showed polymorphisms in studying genotypes, because they showed different banding pattern and discriminated tolerant genotypes from susceptible ones with relation to BINA dhan8 (tolerant) and BINA dhan7 (susceptible) [35]. | |
| Figure 2: SSR Marker Profile of 13 genotypes generated by the primers RM412 and RM336. | |
| Hence parental selection based on the genetic diversity is highly essential to develop a good variety [36]. In future, by utilizing the highly genetic divergent parents of ADT(R) 47 and Nonabokra may help to develop high yielding salt tolerant varieties. The key markers viz., RM412, RM493, RM 3412, RM336, RM17, RM10825, RM148 and RM 206 will be more useful for discriminating the salt tolerant and the salt susceptible genotypes [37]. | |
Acknowledgments |
|
| The work was carried out at Department of Plant Breeding and Genetics, Tamil Nadu Rice Research Institute, Aduthurai during March 2014 to April 2015. The authors are grateful to the DST-SERB PROJECT for funding and acknowledge to finish the work successfully. | |
References |
|
|
|