Research Article, Vegetos Vol: 29 Issue: 4
Morphological and Molecular based Characterization of different Thermo-sensitive Genetic Male Sterile (TGMS) lines in Rice (Oryza sativa L.)
| Pardeep Kumar*, Nautiyal MK, Pankaj Kumar and Kuldeep Kumar | |
| Department of Genetics and Plant Breeding, Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, India | |
| Corresponding author : Pardeep Kumar Department of Genetics and Plant Breeding, Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, India E-mail: pardeepkumar656@gmail.com |
|
| Received: August 31, 2016 Accepted: September 30, 2016 Published: September 30, 2016 | |
| Citation: Kumar P, Nautiyal MK, Kumar P, Kumar K (2016) Morphological and Molecular based Characterization of different Thermo-sensitive Genetic Male Sterile (TGMS) lines in Rice (Oryza sativa L.). Vegetos 29:4. doi: 10.5958/2229-4473.2016.00101.4 |
Abstract
Morphological and Molecular based Characterization of different Thermo-sensitive Genetic Male Sterile (TGMS) lines in Rice (Oryza sativa L.) Pardeep Kumar*, Nautiyal MK, Pankaj.
The present investigation was carried out at the Norman E. Borlaug Crop Research Center of Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Uttarakhand (India) during 2013 and 2014. The eighteen TGMS lines were characterized based on molecular and morphological data. Result on morphological and molecular characterization of TGMS lines showed that, not a single line was observed good for all the traits, different lines good for different traits. Among the eighteen TGMS lines ten lines viz, TGMS-2, TGMS-3, TGMS-4, TGMS-5, TGMS-7, TGMS-8, TGMS-10, TGMS-11, TGMS-14 and TGMS-15 showed complete sterility at average temperature 28.53 to 28.92°C. Based on the molecular genetic diversity data indicated that total of 47 alleles were amplified using sixteen SSR markers in the present study of eighteen TGMS lines all were found polymorphic except for one marker. The range of alleles was 2 - 5, while the average number of alleles per primer was 2.93 and thirteen rare alleles found in different lines. The polymorphic information content (PIC) for these sixteen SSR markers ranged from 0.1780 to 0.3750 with mean value 0.2543. The range of Jaccard’s similarity coefficient was found to vary from 0.46 (TGMS-7 and TGMS-16) to 0.97 (TGMS-11 and TGMS-12). The UPGMA based dendogram constructed using Jaccard’s similarity coefficient of SSR marker data divided eighteen lines into three clusters. Association study between the banding pattern of different markers and spikeletes fertility/sterility of TGMS lines showed that four lines out of eighteen were fertile. These fertile lines separated to other sterile lines by three markers with unique bands. TGMS-6 and TGMS- 9 showed 200bp specific band by RM 324 marker, TGMS-1 and TGMS-18 showed 180bp and 200bp unique bands and in TGMS-1 also showed 180bp unique band with RM 254 marker, it indicated that the unique bands 180bp and 200bp generated by different markers in different lines responsible for fertility. This molecular diversity analysis may be useful for identification of TGMS lines or may be used for marker assisted selection (MAS) for two line hybrid development.
Keywords: Genetic Male Sterile (TGMS) lines; Rice; RNA; DNA; SSR primers
Keywords |
|
| Genetic Male Sterile (TGMS) lines; Rice; RNA; DNA; SSR primers | |
Introduction |
|
| Rice (Oryza sativa L.) is belonging to the family of grasses, gramineae (Poaceae). In fact, the hybrid rice research was initiated in 1964 [1]. The genetic tools essential for breeding hybrid rice varieties are as the male sterile line (A-line), maintainer line (B-line) and restorer line (R-line) were developed during 1973 [2]. The breeding methodology involves the three approaches (a) Three line method or CMS system which is possible and has been found to be most effective genetic tool for developing hybrids, (b) Two line method or TGMS, PGMS and PTGMS system which is governed by environment and (c) One line system or apomictic system which would enable farmers to use their own seed for the successive crops without experiencing genetic segregation. However, the evolution and wide application of these latter two innovative approaches are not likely to occur in future. Among these, three line approaches is being widely adopted in India and had fruitful resulted in the development of more than thirty five varieties of rice hybrids. The CGMS is essentially CMS with a provision of fertility restoration by nuclear gene(s). Hence, it is also referred to as CMS system. The role of cytoplasm in causing male sterility in rice was reported back in the fifties and the first usable cytoplasmic male sterility-fertility restoration system in rice was developed by substituting genes of japonica variety-Taichung 65 into the cytoplasm of the indica variety Chinsurah Boro II [3]. However, this could not be exploited for commercial hybrid seed production probably due to strict self- pollinating nature of the crop. The first commercially usable CMS line was developed in China during 1973 from spontaneous male sterile plant isolated in a population of the wild rice Oryza sativa of spontanea on Hainen Island. Discovery of the source, designated as wild abortive (WA) type is considered a landmark in the history of hybrid rice. The success of hybrid rice is based on the important findings of some key genes. The wild abortive cytoplasmic male sterility (WA - cms) was found in a spontaneous mutant of wild rice Oryza sativa f. spontanea in 1970 [4]. Its maintainer gene was present in most of the Chinese varieties and restorer gene in most IRRI varieties. These findings led to the development of the first batch of three line hybrids of rice. The WA – cms has become the most employed system in development of rice hybrids [5,6]. | |
| The important tool for hybrid seed production is Environmentsensitive genic male sterility system, controlled by nuclear gene expression, which is influenced by environmental factors such as temperature, day-length, or both. This male sterility system was first observed in pepper by Martin and Crawford in 1951 and subsequently in different crops. Advantages of the EGMS system, there is no need for a maintainer line for seed multiplication, thus making seed production simpler and more cost-effective. Any fertile line can be used as a pollen parent (PP); therefore, the frequency of heterotic hybrids is higher among two-line hybrids than among three-line hybrids, thereby increasing hybrid breeding efficiency. Negative effects of sterility-inducing cytoplasm are not encountered. The EGMS trait is governed by major genes, thus enabling their easy transfer to any genetic background and thus increasing diversity among the female (EGMS) parents which helps in reducing potential genetic vulnerability among the hybrids. Since there is no need for restorer genes in the male parents of two-line hybrids, this system is ideal for developing indica/japonica hybrids because most japonica lines do not possess restorer genes. | |
| The discovery in 1973 of Nongken 58 S, a PGMS/TGMS japonica rice line [7], provided the first genetic source for the development of two-line system in hybrid rice. The major feature of such PGMS/ TGMS lines is that, under longer day length and higher temperatures they show complete pollen sterility, under the condition they can be used for hybrid seed production, while under shorter day length and moderate temperatures they show almost normal fertility, this period can be used for their multiplication. In 1987, China initiated a collaborative research project involving the exploitation of PGMS/ TGMS lines to develop two-line system rice hybrids. | |
| Among the various types of molecular markers available, microsatellite have recently received greater attention, especially for breeding purposes. Microsatellite markers, also known as simple sequence repeats or SSRs [8,9] are clusters of short (usually 2 to 6) tandem repeated nucleotide bases distributed throughout the genome. Microsatellite markers are in general co-dominant, multiallelic, and highly polymorphic genetic markers. Microsatellite allele typing requires small amounts of DNA for straightforward PCR and gel electrophoresis analysis [10]. Its main disadvantage is the high cost of the initial investment necessary for marker development. However, the number of microsatellite markers available for a model species such as rice is high, and advantage can be taken of this technology in rice genetic research. Hundreds of microsatellite markers have been physically mapped in the rice genome and can potentially be used, as anchor markers, for comparative genetics, trait mapping and gene isolation. Oryza sativa is composed of two major subspecies, Indica and Japonica (both tropical and temperate) and several ecotypes. Several efforts have been made to assess the genetic diversity within Oryza sativa at both phenotypic and molecular levels. To estimate genetic diversity among Oryza species, several types of molecular markers, particularly simple sequence repeats (SSR), have been used [11-17]. Polymorphisms in the SSR region are considered the results of different replications of repeated sequences, resulting in different sizes of the PCR products. However, alleles with different sequences but having the same length may yield ambiguous results of the phylogenetic analysis. Sequencing SSR products can provide clear information on the evolutionary history of these loci [18,19]. | |
Materials and Methods |
|
| The present investigation was carried out at the Norman E. Borlaug Crop Research Center of Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Uttarakhand (India) during 2013 and 2014. The eighteen TGMS lines were characterized based on molecular and morphological data (Table 1). | |
| Table 1: List of eighteen TGMS lines and their parentage. | |
| Different characters observed for the morphological characterization viz. days to 50% flowering, plant height, number of tillers per plant, number of spikelets, panicle type, panicle length, apiculous pigmentation, stigma color, awning, anthesis time, anthesis time duration (min.), glume angle, panicle exertion %, stigma exertion %, stem color and pollen sterility. The some formulas used for the observing of some traits are: | |
 |
|
| Molecular characterization done by SSR (simple sequence repeats) markers and CTAB procedure was used for isolation of DNA [20]. The quantification of genomic DNA was done by taking the absorbance on Genesys (Table 2) UV spectrophotometer. The optical density was measured at 260 nm and 280 nm. The concentration of the DNA in the sample is related to optical density by the following formula: | |
| Table 2: List of SSR marker used for characterization of TGMS lines. | |
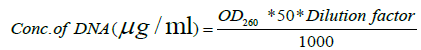 |
|
| The ratio of OD260/280 is an indication of the amount of RNA or protein contamination in the preparation. A value of 1.8 is optimum for best DNA preparation. A value of the ratio below 1.8 indicates the presence of protein in the preparation and a value above 1.8 indicates that the sample has RNA contamination. | |
| PCR reactions were carried out in 9.4 μl PCR master mix and 2 μl genomic DNA. Different SSR primers were taken for amplification. Tdw=6.8 μl, Buffer=1 μl, dNTPs=0.40 μl, Taq Polymerase=0.20 μl and Primer=1 μl boh for Forward and Reverse primer. The PCR reaction was setup for first cycle on 94°C for 5 minutes, second step for 35 cycles on 94°C for one minute for denaturation, 45 seconds for annealing temperature and 72°C for two minute for polymerization and last step 72°C for five minutes. PCR products thus obtained were fractionated by using horizontal gel electrophoresis assembly by agarose gel. Agarose gel of 2.5% concentration for SSR primers were prepared by dissolving the calculated amount of agarose in 10 X TBE buffer [21] and which were examined under the gel-documentation instrument. Gels will be documented using Gel Doc system (Bio-Rad). Data were scored as 1 (present) and 0 (absent) for all the alleles of each of the SSR locus. Polymorphism information content was computed as | |
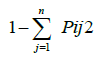 |
|
| Where, | |
| Pij is the frequency of the jth allele at the ith locus summed over the number of alleles (n). Pair-wise similarity and cluster analysis was done using computer software (NTSYS) was used to perform the similarity matrix analysis using ‘UPGMA’ with Jaccard’s coefficient of similarity. | |
Result and Discussion |
|
| The eighteen TGMS line were evaluated for sixteen different traits and based on these traits the line were identified as best line for hybrid seed production. The observations recorded on the floral and morphological characters of TGMS lines are listed in Table 3. | |
| Table 3: Floral and morphological traits recorded in TGMS lines. | |
| For days to flowering 12 lines significant and had mean value high. The early flowering lines were UPRI-97-60-8 (96), UPRI-99-74- 3 (108) and UPRI-99-70-1 (110). For plant height 11 promising lines had significantly and high mean values. Lines with dwarf plant height were UPRI-99-71-1 (72.30), UPRI-99-73-4 (76.31) and UPRI-99-73- 3 (76.54). For number of effective tillers, with more tillers number lines were UPRI-99-74-4 (15.1), UPRI-99-78-1 (14.5) and UPRI-99- 72-4 (14.2). Two types of stem color observed were green and purple, most of the lines showed purple stem color above the ground stem and seven lines showing the green stem color. Intermediate and open type panicles were observed in these 18 lines. All the lines produced intermediate panicle type except for six lines. For panicle, the promising lines which had maximum length of panicle were UPRI- 99-72-3 (25.28), UPRI-99-72-1 (24.10) and UPRI-99-74-1 (24.04). The number of spikeletes varied from minimum in UPRI-99-73-3 (127.6) to maximum in UPRI-99-78-1 (303.5). Eight lines had the significantly higher mean value. Based on the presence and absence TGMS lines divides in to two categories, all the lines have absence of awns but five lines with awns. The color of Apiculous varied from purple to absent, most of the lines having purple apiculous color and seven lines have not any color. The purple Apiculous and color can be used as a morphological marker for identification of these lines. The color of stigma varied from purple to white, only three lines have the white color stigmas viz. UPRI-99-71-1, UPRI-99-73-1 and UPRI-99-73-2. | |
| The glume angle varied from 18° (UPRI-99-74-4) to 33° (UPRI- 99-73-1). Nine lines showing significantly high mean value. The lines which having highest glume angle viz, UPRI-99-73-1 (33.0), UPRI-99-74-3 (28.4), UPRI-99-72-1 (28.2), UPRI-99-74-1 (27.2) and UPRI-99-73-4 (27.0). Thiagarajan et al. reported the glume angle range from 15° to 25°, Singh and Rang) reported the range 35° to 48° and Ravneet S. Behta et al. reported the range of 23.43° to 30.20°. For panicle exertion per-cent varied from 69.73 % (UPRI-99-71-1) to 84.10 % (UPRI-99-71-2). The ten lines showing the significantly high mean value for panicle exertion percentage out of ten, three lines with higher percentage of panicle exertion viz. UPRI-99-71-2 (84.10), UPRI-99-73-1 (82.82) and UPRI-99-73-2 (82.43). For stigma exertion percentage the nine lines were showing significantly high mean value. The lines were with highest percentage of exserted stigma viz. UPRI-99-72-1 (80.05), UPRI-99-78-1 (79.50) and UPRI-99-73-1 (77.19). The anthesis time starts from 8:00 and end to 12:30 for all the lines. For anthesis time duration long is good for pollination and hybridization for production of hybrid. The promising lines are which have long duration for anthesis viz. UPRI-99-74-3 (205), UPRI-99- 71-2 (185) and UPRI-99-72-1 (180). For Pollen sterility (Figure 1) study showed that the mean of sterility was 85%. The range of pollen sterility percentage was 44.32 % (UPRI-97-60-8) to 98.84 % (UPRI- 99-73-1). Therefore lines showed high value over mean. The lines are which showed maximum sterility in the field viz. UPRI-99-73-1 (98.84), UPRI-99-73-2 (97.64), UPRI-99-79-1 (96.75), UPRI-99-72-3 (96.64) and UPRI-97-60-1 (92.38). | |
| Figure 1: Microscopic pollen sterility study of different TGMS lines of rice. | |
| For characterization of different TGMS lines based on floral and morphological traits, different researchers found same findings for different traits with high mean value viz. Salgotra et al. and Thiyagarajan et al., studied the effective tiller number per plant, panicle exertion, number of spikeletes, glume angle, pollen sterility Ramakrishna et al. and Singh et al., plant height, panicle type, panicle exertion, stigma exertion, Apiculous pigmentation, stigma color, time of anthesis, pollen sterility, duration of anthesis and glume angle used for characterization of different lines. Floral biology and morphological characterization of TGMS lines Celine et al. studied the stigma color, pollen sterility, apiculous color and panicle type. Kavithamani et al. studied the traits viz. stigma exertion, days to flowering, pollen sterility and stigma color. | |
| The different researchers have reported the average and critical temperature for different TGMS lines Lohithaswa et al. found 35/23°C and Viraktamath & Virmani found 32/24°C temperature for sterility. Sanchez and Virmani did correlation analyses between spikelet fertility and maximum and daily mean temperatures up to days before heading to determine the critical stage where temperature influences the sterility/fertility expression of the TGMS lines and found that 32°C temperature for sterility. Ramkrishna S et al., and Latha and Thiyagarajan found >30°C temperature for sterility. Salgotra et al. and Celine et al., found 35.4°C critical temperature for sterility. | |
| Therefore among the eighteen TGMS lines ten lines viz, TGMS-2, TGMS-3, TGMS-4, TGMS-5, TGMS-7, TGMS-8, TGMS-10, TGMS- 11, TGMS-14 and TGMS-15 showed complete sterility at critical temperature 28.53 to 28.92°C. These can be used in two line hybrid development. | |
| Out of eighteen lines four lines viz. TGMS-1, TGMS-6, TGMS-9 and TGMS-18 showed fertile spikelets in both main season during 2013 and 2014 but showed complete sterile spikelet in 2014 offseason. It indicated that these lines required high temperature compare to main season because in offseason the average temperatures for these lines were 31.74 to 31.94°C, so these lines required >29°C critical temperature for complete sterility Table 4. | |
| Table 4: Critical sterility temperature for different TGMS lines. | |
| Another four lines viz. TGMS-12, TGMS-13, TGMS-16 and TGMS-17 found that seed setting in kharif, 2013 and complete sterility in kharif and offseason 2014. It also indicated that the critical sterility temperature for these lines slightly higher 29.39-29.53°C than the 2013 kharif season. | |
| Therefore, in Pantnagar situation to get complete sterility these lines should be sown in the last week of May and transplanting be done after 25 to 30 days of seedlings for two line hybrid programme would be appropriate. | |
Molecular Characterization of TGMS Lines |
|
| The 18 TGMS lines of rice namely; UPRI-99-70-1, UPRI99-71- 1, UPRI-99-71-2, UPRI-99-73-1, UPRI-99-73-2, UPRI-99-73-3, UPRI-99-73-4, UPRI-99-74-3, UPRI-99-79-1, UPRI-97-60-1, UPRI- 99-72-1, UPRI-99-72-3, UPRI-99-72-4, UPRI-99-74-1, UPRI-99- 74-3, UPRI-99-75-1, UPRI-99-78-1 and UPRI-97-60-8 were taken for diversity analysis using sixteen simple sequence repeats (SSRs) primers. The overall results of SSRs analysis have been presented in Table 5. All sixteen primers gave clear and consistent amplification and amplified products were ultimately used for DNA profiling. | |
| Table 5: Range of SSR loci scored, number and size of exclusive loci amplified in the Rice genotypes. | |
| Jaccard’s similarity coefficients among 18 TGMS lines of rice namely; UPRI-99-70-1 (1), UPRI99-71-1 (2), UPRI-99-71-2 (3), UPRI-99-73-1 (4), UPRI-99-73-2 (5), UPRI-99-73-3 (6), UPRI-99- 73-4 (7), UPRI-99-74-3 (8), UPRI-99-79-1 (9), UPRI-97-60-1 (10), UPRI-99-72-1 (11), UPRI-99-72-3 (12), UPRI-99-72-4 (13), UPRI- 99-74-1 (14), UPRI-99-74-3 (15), UPRI-99-75-1 (16), UPRI-99-78- 1 (17) and UPRI-97-60-8 (18) are presented in Table 6. Jaccard’s similarity between the pair of genotype varied from maximum 0.97 between UPRI-99-72-1 (11) and UPRI-99-72-3 (12) to a minimum 0.46 between UPRI-99-73-4 (7) and UPRI-99-75-1 (16). Among the eighteen lines the three pairs with lowest genetic similarity (GS) value i.e., maximum diverse pairs were UPRI-99-73-4 (7) and UPRI-99-75- 1 (16) (46% genetic similarity), UPRI-99-73-3 (6) and UPRI-99-71-2 (3) and UPRI-97-60-8 (18) & UPRI-99-73-2 (5) (with GS value 48%). Similarly, three pairs with maximum GS value i.e., minimum diversity in the experimental material of the present study were UPRI-99-72-1 (11) and UPRI-99-72-3 (12) (GS value 97%), UPRI-99-72-4 (13) and UPRI-99-72-1 (11) (GS value 95%) and between UPRI-99-72-4 (13) and UPRI-99-72-3 (12) (GS value 93%). | |
| Table 6: Similarity coefficient between genotypes using sixteen SSR primers. | |
| The objective of the experiment was to estimate the level of genetic diversity among the TGMS lines of rice using 16 SSRs markers. The UPGMA dendrogram was constructed using Jaccard’s similarity coefficients based on SSRs markers data generated on 18 TGMS lines (Figure 2). A total of 47 alleles were amplified using 16 SSR primer pairs in the present analysis. All the lines except for marker RM499 were found to be polymorphic. The range of alleles was 2-5, while the average number of alleles per primer was 2.93. Shah et al. observed 5 to 17 alleles per locus with a mean of 9.4 alleles per locus. | |
| Figure 2: SSR profile of eighteen rice TGMS lines with RM154, RM190 and RM276 markers. | |
| The dendrogram constructed from SSR data divided eighteen TGMS lines in to three groups, I, II and III (Figure 3). | |
| Figure 3: Dendrogram constructed of 18 TGMS lines. | |
| The cluster I was formed at 0.59 Jaccard’s coefficient of similarity. It consisted of two genotypes [UPRI-99-73-3 (6) and UPRI-99-73-4 (7)] clustered together at similarity coefficient 0.74. Group I related to Group II at similarity coefficient of 0.61. | |
| Cluster II consisted of two sub-clusters; sub-cluster IIa and subcluster IIb. Sub-cluster IIa contained ten genotypes. Of these, UPRI- 99-72-1 (11) and UPRI-99-72-3 (12) were not further separated indicating the high level of genetic similarity (0.97) between the two, i.e., some ancestral relationship seems to be there between UPRI-99- 72-1 (11) and UPRI-99-72-3 (12). | |
| Sub-cluster IIb consisted of only one genotypes UPRI-99-75-1 (16) which was related with sub-cluster IIa by similarity coefficient of 0.77. UPRI-99-75-1 (16) line was diverse from the other lines. Cluster III consisted of five genotypes among these two lines UPRI- 99-73-1 (4) and UPRI-99-73-2 (5) which were related to each other by similarity coefficient of 0.57. The maximum similarity coefficient observed between UPRI99-71-1 (2) and UPRI-99-71-2 (3) was 0.76. Cluster III related to cluster II at 0.65 similarity coefficient. | |
| Association study between the banding pattern of different markers and spikeletes fertility/sterility of TGMS lines (Table 7) showed that four lines among the eighteen were fertile. All the three clusters contained one or two fertile lines in each namely, cluster I (TGMS-6), cluster II (TGMS-9) and (TGMS-18) and cluster III (TGMS-1). These fertile lines separated to other sterile lines by three markers with unique bands. TGMS-6 and TGMS-9 showed 200bp specific band by RM 324 marker, TGMS-1 and TGMS-18 showed 180bp and 200bp specific bands and in TGMS-1 also showed 180bp specific band with RM 254 marker, it means that the specific bands 180bp and 200bp generated by different markers in different lines responsible for fertility. This molecular diversity analysis may be useful for identification of TGMS lines. Another association between marker and TGMS lines, study for gca of TGMS lines for number of traits, there were four TGMS lines good for different traits viz, TGMS- 14, TGMS-17, TGMS-10 and TGMS-18. Among these lines only two lines showed specific band only by one marker that were TGMS-10 and TGMS-17 with 150bp specific band by RM 276 marker, this may be helpful for identification of good TGMS lines for number of traits and used in hybrid development for getting higher yield. | |
| Table 7: Association of TGMS lines fertility/sterility with molecular marker data. | |
References |
|
|
|
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi