Review Article, J Plant Physiol Pathol Vol: 9 Issue: 7
Guide for Designing Species Specific Primer
Charan Nadipineni*
Assistant Professor, Department of Genetics, India
- *Corresponding Author:
- Charan Nadipineni
Assistant Professor, Department of Genetics, India
Tel: +91 8217205969
E-mail: n.charan97@gmail.com
Received Date: June 27, 2021; Accepted Date: July 23, 2021; Published Date: July 30, 2021
Citation: Nadipineni C (2021) Guide for Designing Species Specific Primer. J Plant Physiol Pathol 9:7. 256.
Copyright: © All articles published in Journal of Plant Physiology & Pathology are the property of SciTechnol, and is protected by copyright laws. Copyright © 2020, SciTechnol, All Rights Reserved.
Abstract
To identify, characterize and differentiate the pathogen strains, a PCR-based technique is developed. In this method, speciesspecific primers need to be designed with which we can know whether a particular pathogen strain is present or not among a group. To design such types of primers, we need sequences of the related species. All these sequences are aligned and a unique SNP that differentiates a particular species from the rest is used for designing a specific primer. A protocol has been developed for this; by following this researchers can easily identify their desired strain and can use it for breeding purposes.
Keywords: Primer, Species, Fusarium, Gene
Introduction
The PCR-based technique is the fast, sensitive, and highly dependable way for the identification of strains at the intraspecific level. This method can also be used for determining the level of diversity which is very important because higher the diversity, rapid the change in genetic structures resulting in the development of more virulent strains. Many articles have been published on this topic, but none of them clearly explained the procedure in designing species-specific primer. This article is a complete guide for designing species-specific primers by using the programs which are freely available on the internet. To design a specific primer for a species, sequences of most of the species of that particular genus should be analyzed. The more the number of species sequence we analyze, the higher will be the specificity of the primer to that particular species. We need to look for a SNP that differentiates specific species from the rest. In most of the research works, people work with species whose sequences are not yet available in the database. In such cases, a conserved primer needs to be designed for a conserved gene (Eg: Gas1 gene, EF-1 alpha gene, and ITS region in fungal pathogens) by using the available sequences from the database. These conserved genes are the genes that are involved in every step of the organism’s evolution without much change in the coding sequence (cds). The exon region of these genes is highly conserved which can be used for common primer designing and a unique SNP of a species is used for specific primer designing. The designed common primer is used to amplify the conserved gene region of the species followed by cloning the PCR amplicons into a vector to reduce the noise during sequencing. These sequences along with the available sequences are used in specific primer designing. This entire procedure is represented in the flow chart and illustrated step-wise by designing a specific primer for one of the Fusarium species. Fusarium is one of the major agricultural pathogens causing high economic damage to the crops. Apart from reducing the yield, the grain quality gets deteriorated due to the contamination from toxic mycotoxins. Fusarium is large a genus of filamentous fungi that includes several species which are plant pathogens [1]. These species have a broad host range. Under field conditions, one can identify the Fusarium infestation only after the appearance of the symptoms. The measurements against the Fusarium at this stage are not efficient, resulting in yield loss. Different Fusarium species have different sensitivity responses to the fungicide’s application [2]. So, it is very important to know which species of Fusarium has infested the plants.
Currently, the identification of Fusarium is based on morphological and physiological characteristics, which is a time-consuming approach and requires expert knowledge, difficult for a non-specialist. These characteristics are not always well developed, they may be absent in some isolates making things more complicated [3]. Considering these facts rapid and accurate identification of Fusarium species is required for the management of the disease at early stages. So, the focus is shifted towards molecular approaches like Polymerase Chain Reaction (PCR) by using species-specific primer, and many research works for designing specific primers for several Fusarium species has already been published. Sequences from these publications are used to show the way for designing specific primers [4-6].
Steps in species specific primer designing
Sequences retrieval from the database: Sequences of few Fusarium species are available in the database. To create species-specific primers, sequences of all the selected species are requisite. For this, we need to design a common primer for a conserved gene by using the available sequences in the database. The gene GAS1 encodes for β-1,3-glucan which is a major structural component of the fungal cell wall that determines hyphal shape and resistance to adverse environmental conditions [7]. This gene is well conserved because of its crucial role in fusarium pathogenesis. The GAS1 gene sequence of Fusarium graminearum is already available in the database. By using this sequence, available sequences of remaining Fusarium species are retrieved from the NCBI database using nucleotide BLAST (change the option to dissimilar sequences) and the accession numbers are tabulated in Table 1. Both coding sequence (CDS) and genomic sequence (gDNA) are required to get the intron data based on which alignment is done. These sequences are downloaded in FASTA format. Unfortunately, the genomic DNA of F.culmorum is not available in the database but the CDS is available [8,9].
| Species | Locus Tag of GAS1 | Accession number |
|---|---|---|
| F.graminearum | FGSG_00006 | HG970332.2 |
| F.oxysporum | FOXG_17504 | XM_108397513.1 |
| F. verticillioides | FVEG_17015 | XM_018906256.1 |
| F. culmorum | None | LT598659.1 |
| F. fujikuroi | FFUJ_11202 | HF679033.1 |
Table 1: Locus tags, genomic and CDS accession numbers of Gas1 gene for the five Fusarium species.
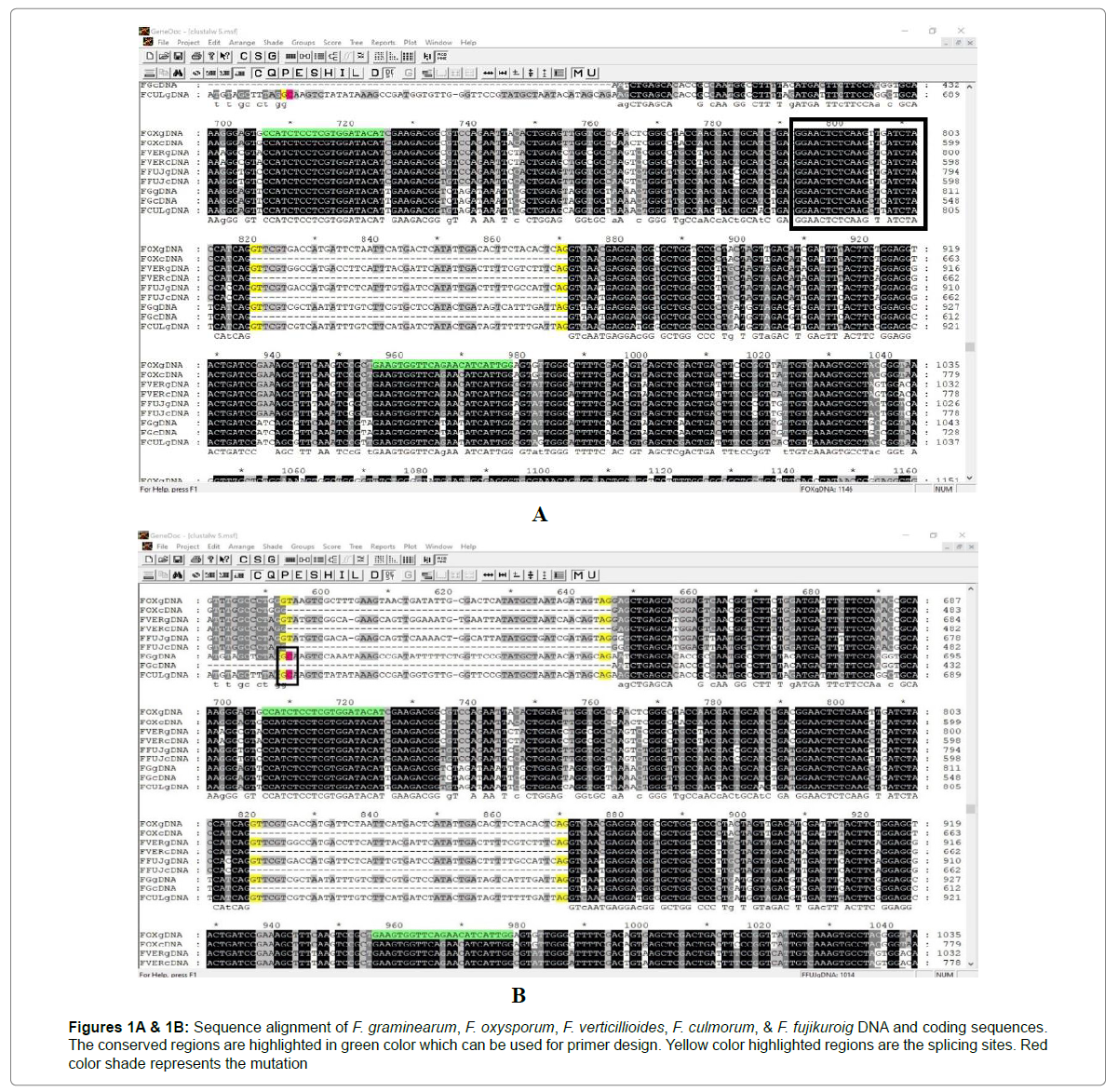
Conserved primer designing: The FASTA format files are uploaded in the CLUSTALW tool (available on the internet) for aligning the sequences. After alignment, the alignment data is downloaded in MSF format. The obtained file in MSF format is opened in GENEDOC (A freely available application) for common primer designing. Intron always consists of two distinct nucleotides at either end: GT at the 5’ end and AG at the 3’ end. These nucleotides are part of the splicing sites. The alignment that we got from the CLUSTALW tool is corrected based on this GT-AG rule of introns. The F. graminearum sequence is a lot similar to the F. culmorum and both of these species have one exon less in comparison to the remaining Fusarium species because of a mutation at the splicing site of F. culmorum and F. graminearum (marked in red color in Figure 1B) where GC is there instead of GT. This mutation results in loss of one exon which might affect the gene function or led to the formation of new protein due to the exon skipping during alternative splicing which may have a new function. If the mutation results in functional loss, we can avoid this by Ethyl Methane Sulfonate (EMS) treatment. EMS chemically modifies the nucleotides resulting in mispairing and base changes. The majority (99%) of the time, EMS induces C-to-T changes resulting in C/G to T/A substitutions. By EMS treatment the mutation “C” changes to “T” resulting in active splicing site GT. After correcting the alignment, the common primer is designed by looking for the highly conserved region across those 10 sequences [10].
To design the best primer, the following rules need to be followed:
1. Strong bond should be there at the first attaching site. That’s why we need to make sure that G/C is present at the 5’ prime end.
2. Optimum GC content is 40-60%. More or less may result in unspecific bands and even the annealing temperature of the primer will depend on this percentage.
3. Optimum length is 18-22bp, less than 18bp may result in unspecific bindings whereas greater than 22bp results in secondary structures.
4. SNP position should always be at the 3’ end of a primer. If SNP is present at the 5’ end, the primer can still attach to the DNA and that SNP remains like an overhang. Secondary structure formations will be there if the SNP position is in the middle.
The sequences CCATCTCCTCGTGGATACAT and GAAGTGGTTCAGAACATCATTGG that are found to be conserved across the 10 sequences are selected as forward and reverse primer respectively for the GAS1 gene as shown in Figures 1A & 1B. The properties of these primers are checked by using OLIGOCALC and tabulated in Table 2. The product size for these primers is 160bp [11-13].
Figures 1A & 1B: Sequence alignment of F. graminearum, F. oxysporum, F. verticillioides, F. culmorum, & F. fujikuroig DNA and coding sequences. The conserved regions are highlighted in green color which can be used for primer design. Yellow color highlighted regions are the splicing sites. Red color shade represents the mutation
| Primer | Sequence | Length | GC% | Annealing temperature (oC) |
|---|---|---|---|---|
| Forward | CCATCTCCTCGTGGATACAT | 20 | 50 | 51.8 |
| Reverse | GAAGTGGTTCAGAACATCATTG | 22 | 45 | 53.5 |
Table 2: Properties of common forward and reverse primers.
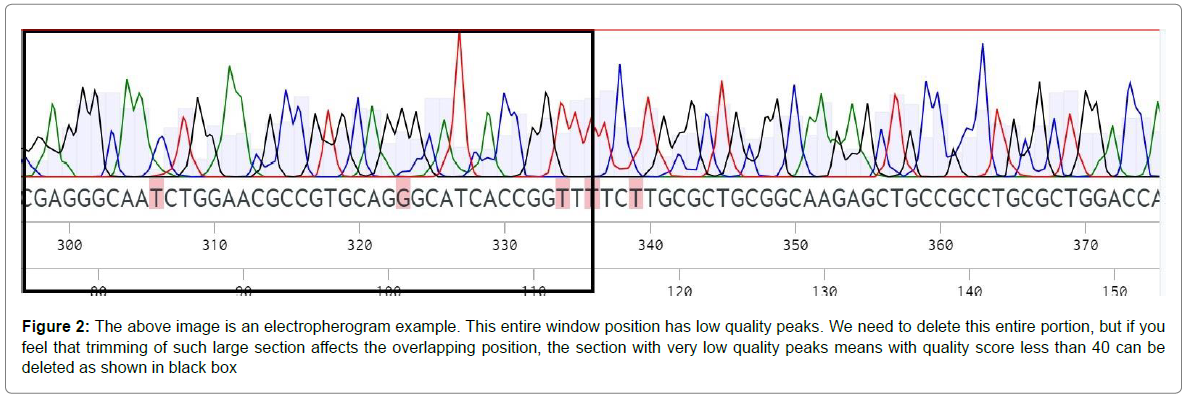
Quality evaluation of sequences extracted from the vector: By using the common primers, the DNA from 15 Fusarium species is amplified separately. We cannot send these PCR amplicons directly for sequencing due to the mistakes made by the polymerase enzyme. To reduce these errors or noise during sequencing, these amplicons are cloned into a vector. Based on my experience, TOPO vector is best for cloning since it’s easy to handle without complications. The plasmid from the positive colonies is extracted and sent for sequencing. The electropherograms (Raw sequencing data) of the 15 Fusarium strains are opened in CHROMAS (freely available application) to select the best sequence data. The general rule is to allow only 1-2% error in the sequence data; higher error rates make the correction more difficult [14-15]. For each Fusarium species, two files will be there: forward and reverse sequence. By using the reverse complement tool, the reverse sequence is reversed and the ends of both these sequences which are of low quality are trimmed (as shown in Figure 2). Then these two sequences are connected after removing the overlapping and the vector sequences. After connecting the sequences, we finally have the Gas1 gene sequences of 15 Fusarium species that can be used for designing species-specific primers. If you want to avoid this entire tiresome work, you can directly upload the raw data in Benchling (an online free tool) and it automatically gives the consensus. This consensus can be used directly for designing species-specific primers.
Figure 2: The above image is an electropherogram example. This entire window position has low quality peaks. We need to delete this entire portion, but if you feel that trimming of such large section affects the overlapping position, the section with very low quality peaks means with quality score less than 40 can be deleted as shown in black box
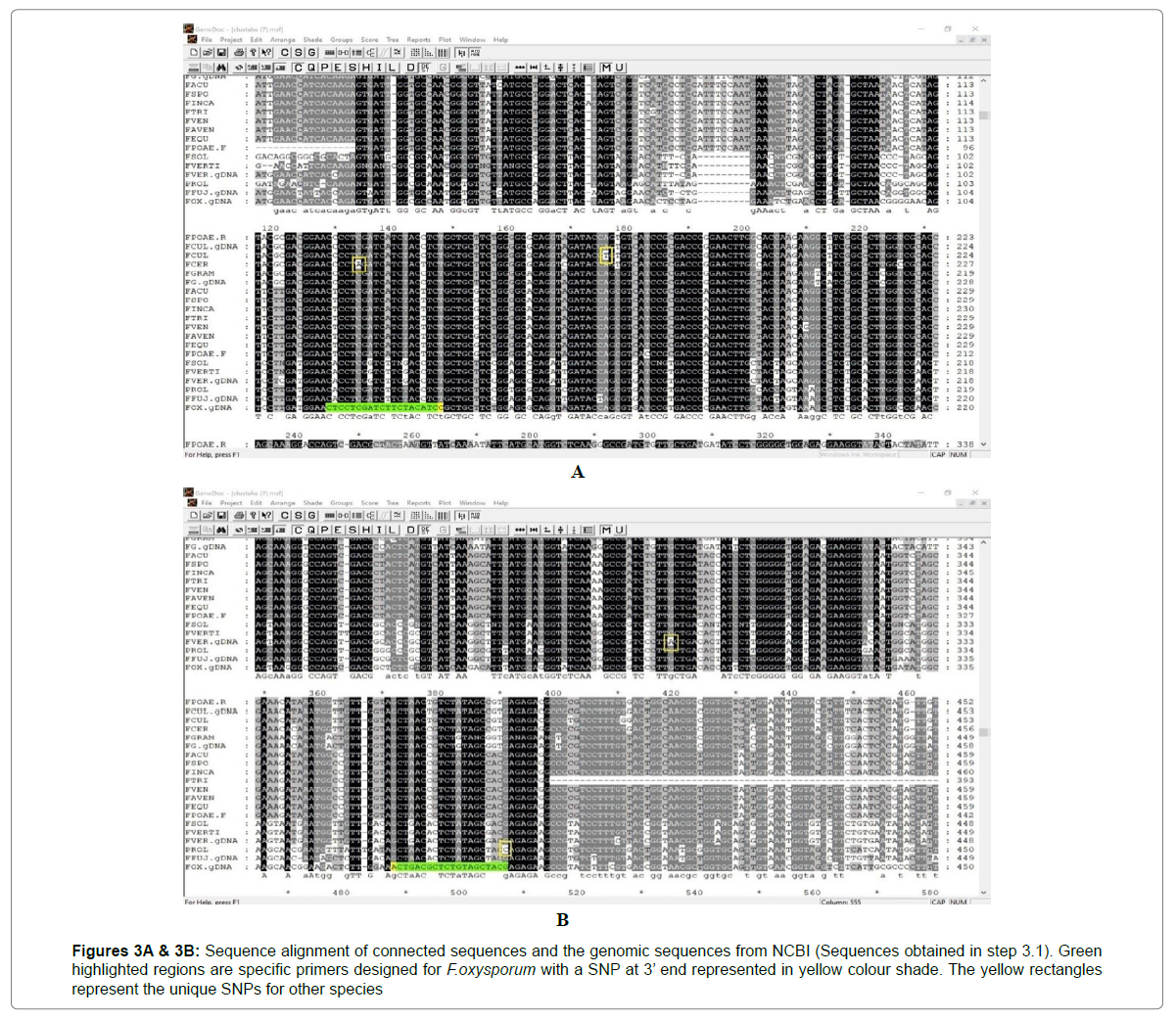
Species specific primer designing: The sequences obtained after cloning are uploaded in GeneDoc and aligned together. Based on this alignment, species-specific primers are designed for each of the Fusarium species. For example, I designed a specific primer for F.oxysporum for the Gas1 gene. The regions with a SNP by which we can differentiate the F.oxysporum from the rest of the species are selected as forward and reverse primers as shown in Figures 3A & 3B and the SNP should be present at the 3’ end. The properties of these primers are checked by using OLIGOCALC and the results are tabulated in Table 3.
| Primer | Sequence | Length | GC% | Annealing temperature (oC) |
|---|---|---|---|---|
| Forward | CTCCTCGATCTTCTACATCC | 20 | 50 | 51.8 |
| Reverse | ACTGACGCTCTGTAGCTACG | 20 | 55 | 53.8 |
Table 3: Properties of species-specific primer for F.oxysporum.
Remaining species also have unique SNPs which can be used for specific primer designing (see Figures 3A & 3B).
Figures 3A & 3B: Sequence alignment of connected sequences and the genomic sequences from NCBI (Sequences obtained in step 3.1). Green highlighted regions are specific primers designed for F.oxysporum with a SNP at 3’ end represented in yellow colour shade. The yellow rectangles represent the unique SNPs for other species
By following these steps, one can easily design the specific primers without complications.
References
- Flood J (2010) The importance of plant health to food security. Food Sec 2: 215-231.
- Chakraborty S, Newton AC (2011) Climate change, plant diseases and food security: an overview. Plant Pathology 60: 2-14.
- Jimenez Garcia (2018) Fusarium mycotoxins and metabolites that modulate their production. 10.5772/intechopen.72874.
- Richard RM, Kris A, Martin K (2018) Control of Fusarium verticillioides (Sacc) Nirenberg and Fumonisins by Using a Combination of Crop Protection Products and Fertilization. Toxins 10: 67.
- TulinAskun (2018) Introductory Chapter: Fusarium: Pathogenicity, Infections, Diseases, Mycotoxins and Management. 10.5772/intechopen.76507.
- Preiser, Viktoria & Goetsch (2014) Development of a multiplex real-time PCR to quantify Fusarium DNA of trichothecene and fumonisin producing strains in maize. Anal Methods 7.
- Kovgaard, Kerstin & Nirenberg (2002) Evolution of Fusarium oxysporum f. sp. vasinfectum Races Inferred from Multigene Genealogies. Phytopathology 91: 1231-1237.
- Akbar A (2018) Detection, virulence and genetic diversity of Fusarium species infecting tomato in Northern Pakistan. PLoS ONE 13.
- Zaira Caracuel (2005) Fusarium oxysporum gas1 Encodes a Putative β-1,3- Glucanosyltransferase Required for Virulence on Tomato Plants. Mol Plant Microbe Interact 18: 11400-1147.
- Gribskov, Michael, Devereux, John (1991) Sequence Analysis Primer.
- Tkacz, Jan S, Lange (2004) Advances in Fungal Biotechnology for Industry, Agriculture and Medicine.
- Hope R, Aldred D, Magan N (2005) Comparison of environmental profiles for growth and deoxynivalenol production by Fusarium culmorum and F. graminearum on wheat grain. Lett Appl Microbiol 40: 295-300.
- Zaira Caracuel (2005) Fusarium oxysporum gas1 Encodes a Putative β-1,3- Glucanosyltransferase required for Virulence on Tomato Plants. Mol Plant Microbe Interact 18: 1140-1147.
- Zhenyue Lin (2014) Species-Specific Detection and Identification of Fusarium Species Complex, the Causal Agent of Sugarcane Pokkah Boeng in China. PLoS ONE 9: e104195.
- Ida Karlsson (2016) Genus-Specific Primers for Study of Fusarium Communities in Field Samples. Appl Environ Microbiol 82: 491-501.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi