Research Article, J Regen Med Vol: 9 Issue: 2
Cortical and Cancellous Bone Regeneration in Cranioplasty and Spinal Arthrodesis Models Using Autologous Homologous Bone Construct (AHBC)
Pratima Labroo*, Nicholas Baetz, Marytheresa Ifediba, Kendall Stauffer, Michael Sieverts, Jessie Johnson, Eric Chan, Ian Robinson, James Miess, Jennifer Irvin, Lyssa Lambert, Caroline Garrett, Edward W. Swanson, and Nikolai A. Sopko
Department of Research and Development, Polarity TE MD, Inc., Salt Lake City, UT, USA
*Corresponding Author: Pratima Labroo
Department of Research and Development, University of Utah, Greater Salt Lake City Area, 84104, United States
Tel: +1 435 512 3176
E-mail: Pratimalabroo@polarityte.com
Received: May 08, 2020 Accepted: June 17, 2020 Published: June 26, 2020
Citation: Labroo P, Baetz N, Ifediba M, Stauffer K, Sieverts M, et al. (2020) Cortical and Cancellous Bone Regeneration in Cranioplasty and Spinal Arthrodesis Models Using Autologous Homologous Bone Construct (AHBC). J Regen Med 9:2. doi: 10.37532/jrgm.2020.9(2).159
Abstract
Autologous bone grafts are commonly used to treat large bone defects. Though autologous bone grafts have high rates of success, they are limited by availability of donor tissue and may not be suitable for all treatment pathologies. Allografts are an attractive alternative as they do not utilize tissue from the patient but have a higher risk of infection and graft failure. In this study, an autologous homologous bone construct (AHBC) derived from viable bone, was compared to autologous bone grafts and demineralized bone matrix in rabbit models of critical-sized cranial defects and spinal fusion. AHBC is made from a small bone harvest obtained from an uninjured area of the patient. Without any exogenous supplementation or culturing, the AHBC is expeditiously deployed to the treatment site, where it initiates osteogenesis and osteoinduction and closes the defect from the inside out with cortico-cancellous bone. Treated defects were assessed using imaging modalities (micro CT, confocal, SEM, multiphoton, Raman spectroscopy), molecular and proteomics analysis, as well as mechanical testing. AHBC performed as well as autograft in all modalities and exceeded autograft in several. Both AHBC and autograft were observed to have more positive outcomes than DBM+BMP2 in both cranioplasty and arthrodesis models. Clinical significance: AHBC was able to regenerate cortical and cancellous bone in cranioplasty and spinal arthrodesis translational models and is a viable alternative to autografts and allografts.
Keywords: Homologous, Osteogenesis, Autologous, Cancellous, Cortical
Introduction
Bone grafts are commonly used for the treatment of criticalsized defects including congenital bone defects and those caused by trauma, cancer, or injury, with autologous bone grafts (ABG) being most commonly utilized [1]. ABGs are harvested from a healthy donor site [2] and transferred to the defect. Advantages of ABGs include avoidance of rejection and communicable diseases and they have improved integration of viable tissue. Complications still exist however, including lack of donor tissue availability, donor site morbidity, infection (at donor and/or recipient site), graft resorption or failure, and loss of bone volume [3]. Allogenic bone products are often used in an attempt to eliminate donor site morbidity, and their use has been steadily increasing [4]. These include intact cadaveric allogenic bone grafts and bone void fillers that are created from processed cadaveric bone compounded with additional chemicals and materials to influence their handling and integration characteristics [5]. Although allografts avoid the need for autologous donor tissue, they have a higher rate of graft failure, treatment site infection, and carry the risk of disease transmission [6]. Demineralized bone matrix (DBM) is the largest class of allogenic bone products, which are available in a variety of configurations [7,8]. DBM retains the collagen component of bone while excluding mineral components and provides a scaffold for growth of osteocytes into the defect [9]. DBM is primarily used as an osteoconductive substrate with limited to no osteoinductive or osteogenic capability [10]. Due to these limitations, DBMs cannot be used alone for criticalsized bone defects [11] and are commonly combined with bone morphogenetic protein 2 (BMP2) to enhance bone regeneration [11]. BMP2 belongs to the transforming growth factor beta (TGF-β) family, which plays an important role in cell propagation and differentiation [12]. BMP2 promotes osteogenesis and chondrogenesis [13,14] however, BMP2 has been known to produce an initial inflammatory response [15], swelling at the surgical site, which can lead to infection and graft rejection [16], and can be oncogenic at high doses [17–19].
An osteogenic autologous homologous bone construct (AHBC) has been developed (Table1). It is produced from a small piece of autologous bone. In this study, AHBC was prepared from a bone harvest containing a cortico-cancellous interface. It is hypothesized that AHBC processing seeks to stimulate an osseous tissue interface and leverage bone tissue’s endogenous regenerative capacity. AHBC is manufactured in a hysiological media void of enzymes and is returned to the defect area and not cultured ex-vivo, utilizing the patient’s body to provide the necessary signaling and support for the AHBC to expand within the defect.
| ID | Hu0080 | Hu0081 | Hu0082 | Hu0084 |
|---|---|---|---|---|
| Bone type | Pubis | Pelvic ramus | Pelvic ramus | Pubis and pelvic ramus |
| Age (years) | 26 | 60 | 48 | 28 |
| Gender | M | M | M | M |
| Source | Donor Connect, Salt Lake City, UT | Donor Connect, Salt Lake City, UT | Donor Connect, Salt Lake City, UT | Donor Connect, Salt Lake City, UT |
| Cause of death | Gunshot wound to head | Unknown cause | Intercranial hemorrhage/Stroke | Asphyxiation |
Table 1: Human donor demographics.
In this study, the effect of AHBC processing and the potential of AHBC as an alternative candidate for cranioplasty and arthrodesis was examined. The transcriptomic and proteomic effect of AHBC processing in human bone tissues demonstrated a priming effect. Treated spinal fusions and critical-sized cranial defects were studied in an established rabbit model and assessed In-vivo using CT, gross imaging, and ex-vivo with μCT, compound microscopy, molecular and proteomic analysis, mechanical testing, Raman spectroscopy, scanning electron microscopy (SEM), and multiphoton microscopy (MP). Use of rabbits for nonclinical toxicity and orthopedic implant evaluations including spinal fusion, cranial and long bone defect repair has been well established as a translational model [20–22]. Preclinical investigations presented herein using this species align with AAMI, ANSI and ASTM industry standards [23–25]. Our findings indicate Rabbit AHBC can induce osteogenesis as seen in the spinal fusion model and regenerate diploic structure as seen in the cranioplasty model with cortical and cancellous bone with density, composition, and tissue architecture analogous to native bone
Methods (additional details in Supplementary methods)
AFM
Topographical and stiffness information of bone was obtained using a WITec Alpha 300RA atomic force microscope (AFM) in Digital Pulsed Force Mode (DPFM).
Viability Assay
Alomar Blue assay was used to evaluate the viability of AHBC and compared to native bone over time.
Cranial defect model
Animals were placed in ventral recumbency for surgery to create two paramedial, 8 mm (outer diameter), bicortical parietal bone defects. A midline skin incision was made from the nasofrontal. The periosteum was incised and reflected away from the underlying parietal bones. Defects were created using a 8 mm short trephine drill and contra angle handpiece on a single implant motor system with continuous isotonic saline irrigation. Animals were randomized using a card-based technique to receive 1) split calvarial autografts (SCA) (n=6), 2) Rabbit AHBC (n=6), or 3) DBM+BMP-2 (n=6). Periosteum was re-opposed over both defects with 4-0 suture (Monocryl®, Ethicon, Inc., Somerville, NJ). AHBC was generated intra-operatively in accordance with manufacturer’s processing methods using the entire quantity of parietal bone harvested from the defect. AHBC manufacturing creates micro aggregates of autologous bone tissue, which retain the endogenous regenerative and support cell populations associated with native bone healing. AHBC processing is designed to optimize the aggregates for passive diffusion and to activate endogenous pathways involved in bone repair. It is in a physiological media void of enzymes or growth factors. It is not cultured ex-vivo, rather the AHBC is placed in the defect following manufacturing
Posterolateral Lumbar Spinal Fusion Model
Animals were placed in ventral recumbency for bilateral iliac crest harvest and transverse lumbar spinous process fusion. Right and left iliac crests and adjacent ilium body were resected with Beyer Rongeurs yielding 1.6 – 1.8 grams of cortico-cancellous bone per crest. AHBC was manufactured using the entire quantity of harvested iliac crest bone. Test article was equally divided between right and left L4-L5 processes to bridge from the cranial aspect of the L4 transverse processes to the caudal aspect of the L5 transverse processes. A dorsal midline skin incision was created from lumbar vertebral body 3 to 6. Bilateral paraspinal incisions were made to expose and reflect the erector spinae muscles and fasciae covering the dorsal and lateral aspects of the right and left L4-L5 transverse processes. A 1.2 mm step drill urban decorticating bur was used to decorticate the dorsal surfaces of the transverse processes of L4 and L5 extending approximately 2 cm laterally from the transverse process (TP)/pars interarticularis junction. Animals were randomized using a card-based technique to receive autologous bone grafts (ABG) (n=6), Rabbit AHBC (n=6), or DBM+BMP-2 (n=6).
AHBC Processing
Necropsy/Macroscopic evaluation
Animals were euthanized on POW (post-operative week) 8 in accordance with the American Veterinary Medical Association (AVMA) Guidelines on Euthanasia [26]. Further details in supplementary methods.
Computed tomography
Longitudinal computed tomography (Vimago™, Epica Medical Innovations, San Clemente, CA, USA) was performed on the day of surgery immediately prior to and following the operation and repeated on post-operative weeks 2, 4, 6, and 8 for all animals in both cranial defect and the spinal fusion studies.
Manual palpation and mechanical testing
Manual palpation test was performed independently at necropsy by two blinded testers as an assessment of spinal fusion and fusion masses as previously described and were given a score of 0 (nofusion), 1 (partial fusion) and 2 (complete fusion) [27]. Load versus displacement and indentation strength testing was performed on explanted cranial defects and spinal fusion masses using an electronic universal testing machine (UTM Instron, Norwood, MA, USA). One indentation location was used on each specimen and 3 indentations were carried out at each location. Load was applied along the transverse axis of the bone.
Raman spectroscopy
Raman microscopy (Raman DXR™ Microscope, Thermo Fisher Scientific, Waltham, MA) was performed and spectral information was collected using Dispersive Raman software (OMNIC™ v.32, Thermo Fisher Scientific, Waltham, MA).
Scanning electron microscopy and multiphoton microscopy
An environmental scanning electron microscope (Zeiss Evo LS 10, Carl Zeiss AG, Oberkochen, Germany) was used for specimen imaging, using a back-scatter detector (High Definition Back- Scatter Detector (HDBSD), Carl Zeiss AG, Oberkochen, Germany. Secondharmonic imaging was performed using a multiphoton confocal microscope (Leica SP8, Leica Microsystems, Wetzlar, Germany) equipped with a 10x 0.40 NA objective
Confocal microscopy
Treated cranial defects were stained with DAPI (Blue-cell nuclei), actin (Red-cytoskeleton) and hydroxyapatite LONZA assay (Greenmineral deposits). Confocal fluorescent imaging was performed using a single photon confocal microscope (Leica TCS SP8, Leica Microsystems, Wetzlar, Germany).
Molecular analysis and proteomic analysis of rabbit and human AHBC
Rabbit and Human native bone and AHBC (n=3) were used for gene expression analysis and proteomic analysis to understand the effect of AHBC processing on native bone specimen. See supplementary materials for additional details. Human AHBC was used due to timely availability of fresh and viable tissue.
Statistical analysis
Data is expressed as mean ± standard deviation unless otherwise noted. Statistics analysis is described in detail in the supplemental material. Additional details regarding sample size and evaluation of primary and secondary outcomes included in supplementary materials.
Results
Evaluation of human and rabbit AHBC Ex-Vivo
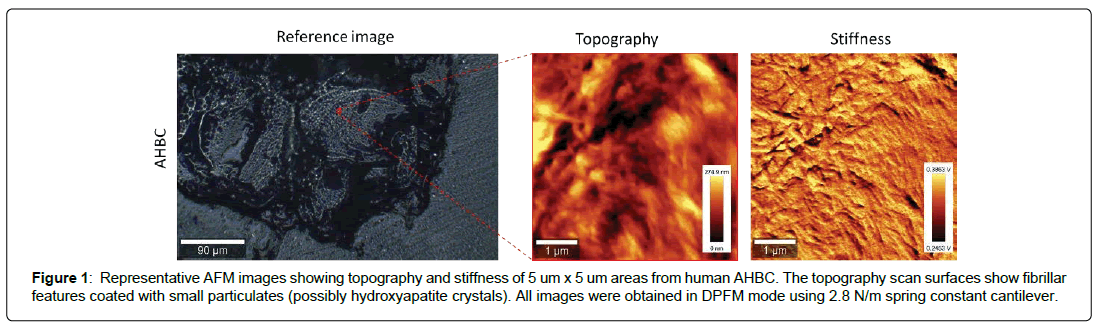
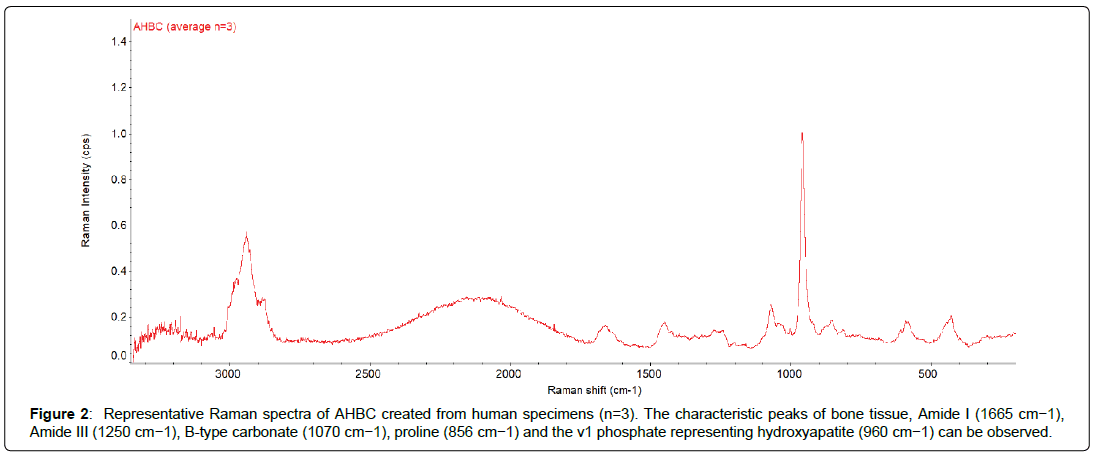
Structural and Chemical Characterization of AHBC: The ultrastructure of AHBC created from fresh cadaveric human bone was determined by AFM. AFM provides the highest resolution amongst all the microscopy techniques and has been used to look at the structural detail of biological tissue. AFM height images (surface topography) and force amplitude images (stiffness) were obtained in Digital pulsed force mode in air for AHBC (Figure 1). AHBC exhibits presence of collagen fibril-like features with mineral deposits across the sample. In addition to AFM, Raman spectra were collected at different points across AHBC to look at its chemical fingerprint (Figure 2). Raman spectroscopy was performed to characterize the chemical composition of AHBC [28]. Raman spectra were collected from the surface of AHBC. N=3 samples per group corresponding to three human specimens analysed. The standard mineral (hydroxyapatite) peak (~965 cm-1) and collagen (proline) peak (~856 cm-1) was observed in the fingerprint region (1800-500 cm-1) of AHBC samples, shown in (Figure 2) These data demonstrate AHBC retains relevant structural and compositional characteristics of native bone tissue that are important for osteogenesis.
Figure 1: Representative AFM images showing topography and stiffness of 5 um x 5 um areas from human AHBC. The topography scan surfaces show fibrillar features coated with small particulates (possibly hydroxyapatite crystals). All images were obtained in DPFM mode using 2.8 N/m spring constant cantilever.
AHBC-processing associated with biochemical changes in protein and transcript profiles.
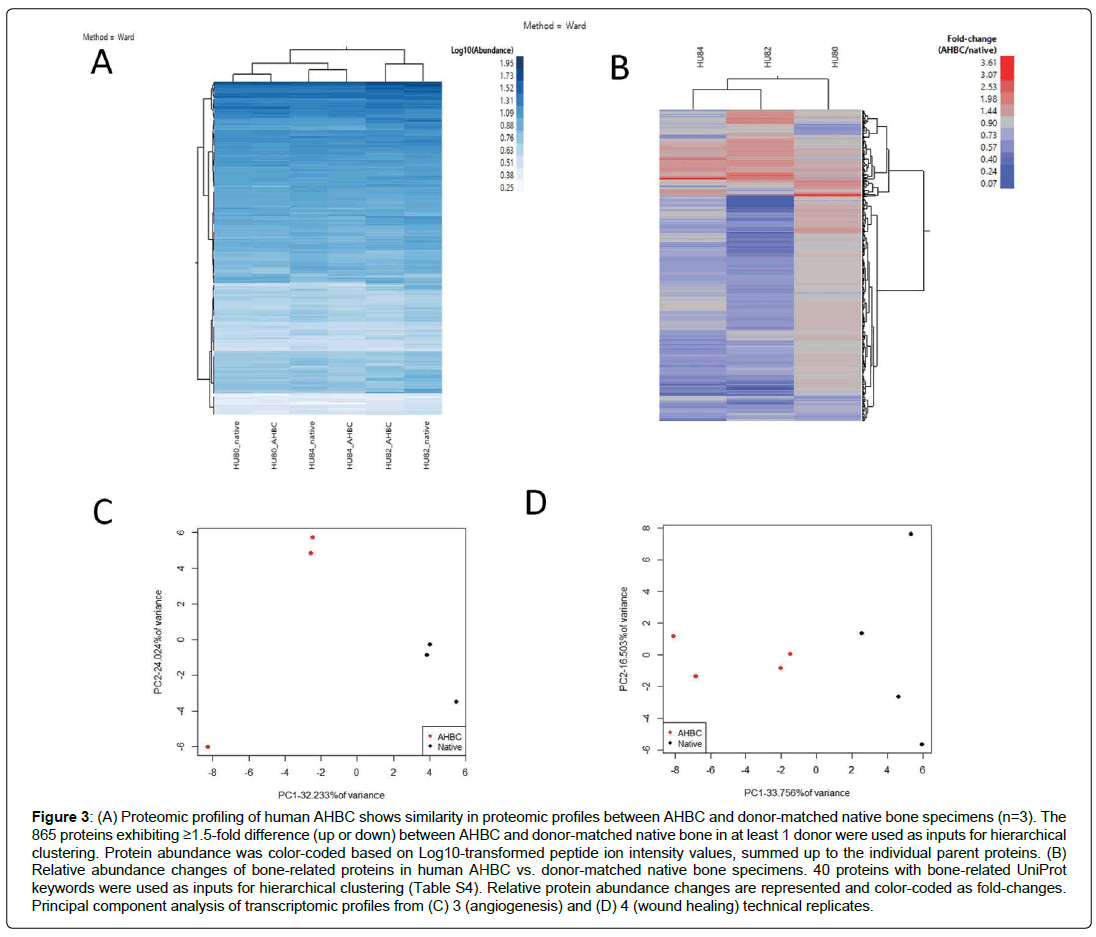
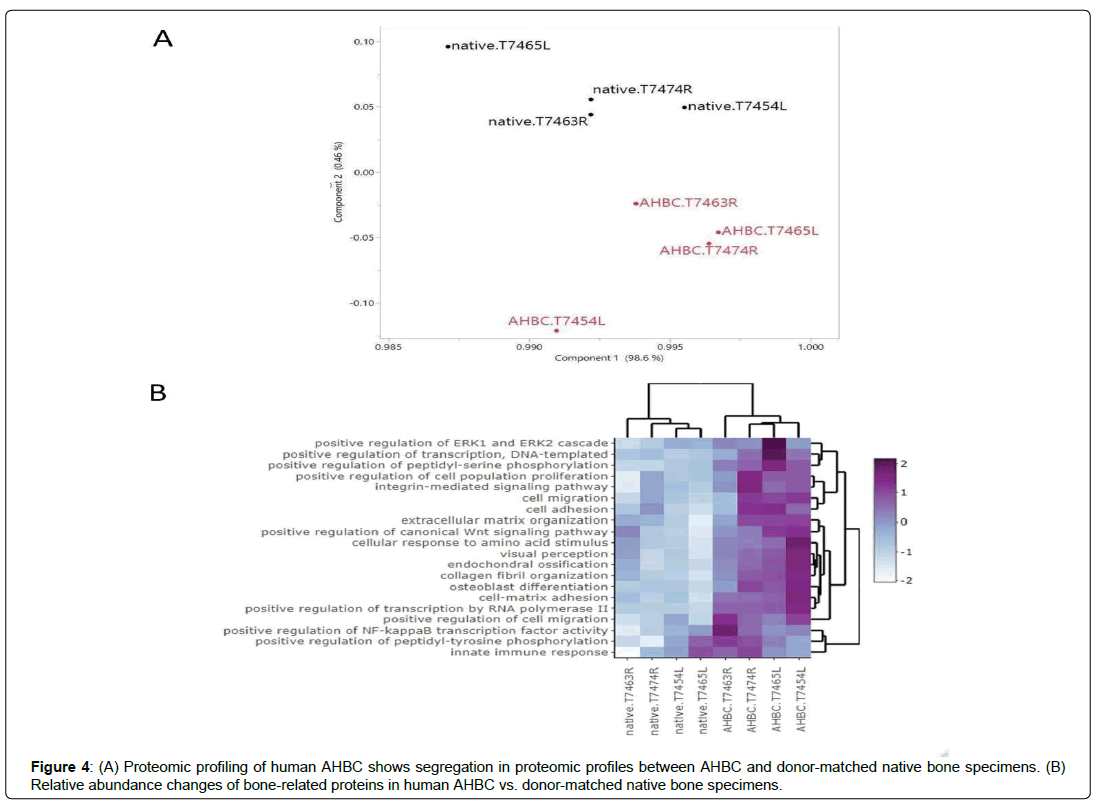
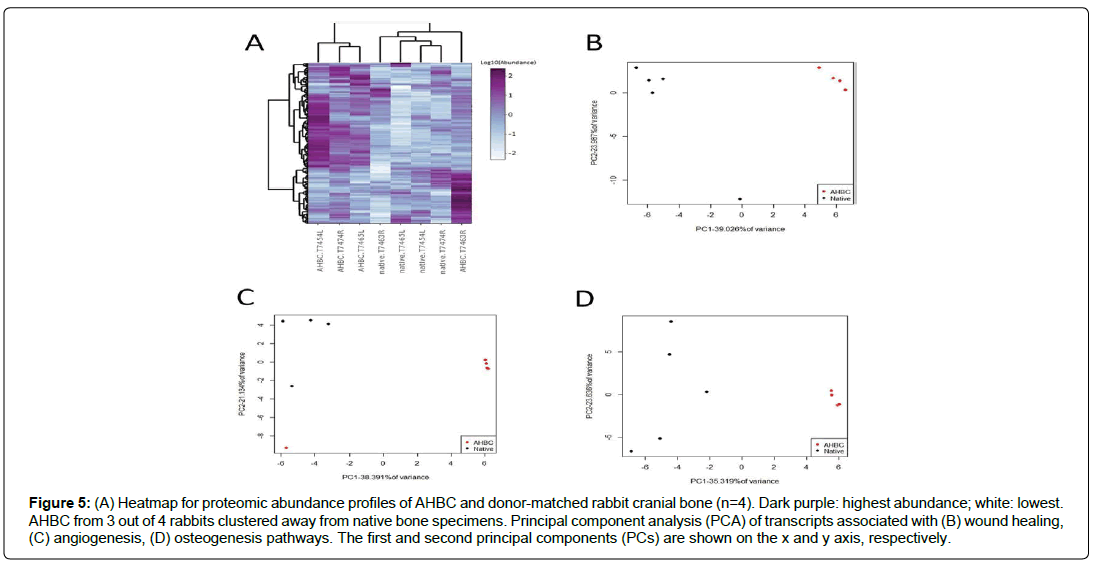
The effect that AHBC processing has on the bone tissues was evaluated. Global proteomic profiles from human cadaveric bone specimens received within 48 hours of death (n=3, Table 1, Donor Connect, Salt Lake City, UT) and the donor-matched AHBC demonstrated similar proteomic profiles between native bone and AHBC (Figure 3A). Targeted assessment of wound healing and angiogenesis pathway transcripts showed differences between native bone and AHBC by principal component analysis (PCA) (Figures 3C & 3D) (Table 2). Similarly, based on gene ontology annotations for the proteomic profiles, protein abundance changes associated with AHBC were significantly enriched for (p<0.01) in osteogenesis-related biological processes, including “extracellular matrix organization” and “negative regulation of TGF-beta receptor signalling” (Table 3). Focusing on bone-related proteins, specific classes of proteins appeared to exhibit concordant abundance changes across the 3 biological replicates. (Figure 3B). The increase in peptidyl-prolyl cistrans isomerases A and B suggested that AHBC-processing was associated with potentiation for bone regeneration, which is associated with collagen synthesis [29] (Table 4) Proteomic profiles of native rabbit cranial bones (n=4) appeared to be different from those of donor-matched AHBC. 741 of the 2735 rabbit proteins quantified showed abundance changes (Table 1), defined as having ≥1.5-fold difference between at least one pair of donor-matched pre- and postprocessing sample. PCA showed a segregation of native vs. AHBC (Figure 4a).
| Term | Count | % | PValue |
|---|---|---|---|
| GO:0034314~Arp2/3 complex-mediated actin nucleation | 8 | 1.24 | 0 |
| GO:0030199~collagen fibril organization | 11 | 1.7 | 0.01 |
| GO:0042593~glucose homeostasis | 6 | 0.93 | 0.02 |
| GO:0030216~keratinocyte differentiation | 6 | 0.93 | 0.02 |
| GO:0051289~protein homotetramerization | 7 | 1.08 | 0.02 |
| GO:0007229~integrin-mediated signaling pathway | 11 | 1.7 | 0.05 |
| GO:0006536~glutamate metabolic process | 4 | 0.62 | 0.06 |
| GO:0002063~chondrocyte development | 4 | 0.62 | 0.06 |
| GO:0006749~glutathione metabolic process | 6 | 0.93 | 0.06 |
| GO:0010595~positive regulation of endothelial cell migration | 6 | 0.93 | 0.06 |
| GO:0045087~innate immune response | 12 | 1.85 | 0.07 |
| GO:0018149~peptide cross-linking | 5 | 0.77 | 0.08 |
| GO:0051897~positive regulation of protein kinase B signaling | 6 | 0.93 | 0.09 |
| GO:0005975~carbohydrate metabolic process | 9 | 1.39 | 0.09 |
Table 2: Functional analysis by GO (biological processes) terms showed an enrichment for processes involved in collagen and structure reorganization.
| Category | Term | Count | % | P Value |
Fold Enrichment | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0030198~extracellular matrix organization | 32 | 3.7 | 0 | 1.75 | 0.98 | 0.98 | 2.07 |
| GOTERM_BP_DIRECT | GO:0030512~negative regulation of transforming growth factor beta receptor signaling pathway | 12 | 1.39 | 0 | 2.68 | 1 | 0.95 | 3.38 |
| GOTERM_BP_DIRECT | GO:0010811~positive regulation of cell-substrate adhesion | 11 | 1.27 | 0 | 2.83 | 1 | 0.88 | 3.58 |
| GOTERM_BP_DIRECT | GO:0045944~positive regulation of transcription from RNA polymerase II promoter | 49 | 5.67 | 0 | 1.48 | 1 | 0.9 | 5.14 |
| GOTERM_BP_DIRECT | GO:0006953~acute-phase response | 11 | 1.27 | 0 | 2.69 | 1 | 0.87 | 5.6 |
| GOTERM_BP_DIRECT | GO:0030239~myofibril assembly | 6 | 0.69 | 0 | 4.41 | 1 | 0.89 | 7.1 |
| GOTERM_BP_DIRECT | GO:0007155~cell adhesion | 44 | 5.09 | 0 | 1.49 | 1 | 0.88 | 7.91 |
| GOTERM_BP_DIRECT | GO:0009611~response to wounding | 9 | 1.04 | 0.01 | 2.89 | 1 | 0.9 | 9.89 |
| GOTERM_BP_DIRECT | GO:0001501~skeletal system development | 16 | 1.85 | 0.01 | 2.06 | 1 | 0.87 | 9.94 |
| GOTERM_BP_DIRECT | GO:0032760~positive regulation of tumor necrosis factor production | 10 | 1.16 | 0.01 | 2.45 | 1 | 0.98 | 19.3 |
| GOTERM_BP_DIRECT | GO:0045087~innate immune response | 39 | 4.51 | 0.01 | 1.45 | 1 | 0.97 | 19.3 |
| GOTERM_BP_DIRECT | GO:0051091~positive regulation of sequence-specific DNA binding transcription factor activity | 11 | 1.27 | 0.01 | 2.26 | 1 | 0.98 | 22.5 |
| GOTERM_BP_DIRECT | GO:0019731~antibacterial humoral response | 8 | 0.93 | 0.02 | 2.74 | 1 | 0.98 | 24.3 |
| GOTERM_BP_DIRECT | GO:0050830~defense response to Gram-positive bacterium | 12 | 1.39 | 0.02 | 2.13 | 1 | 0.97 | 24.9 |
| GOTERM_BP_DIRECT | GO:0006936~muscle contraction | 20 | 2.31 | 0.02 | 1.69 | 1 | 0.98 | 29.3 |
| GOTERM_BP_DIRECT | GO:0031659~positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle | 4 | 0.46 | 0.02 | 5.14 | 1 | 0.99 | 36.8 |
| GOTERM_BP_DIRECT | GO:0050679~positive regulation of epithelial cell proliferation | 7 | 0.81 | 0.03 | 2.77 | 1 | 0.99 | 38 |
| GOTERM_BP_DIRECT | GO:0045214~sarcomere organization | 7 | 0.81 | 0.03 | 2.77 | 1 | 0.99 | 38 |
| GOTERM_BP_DIRECT | GO:0006898~receptor-mediated endocytosis | 20 | 2.31 | 0.03 | 1.63 | 1 | 0.99 | 38.6 |
| GOTERM_BP_DIRECT | GO:0001934~positive regulation of protein phosphorylation | 14 | 1.62 | 0.03 | 1.85 | 1 | 0.99 | 40.2 |
| GOTERM_BP_DIRECT | GO:0050776~regulation of immune response | 15 | 1.74 | 0.03 | 1.79 | 1 | 0.99 | 40.4 |
| GOTERM_BP_DIRECT | GO:0042060~wound healing | 10 | 1.16 | 0.03 | 2.14 | 1 | 0.99 | 42.5 |
| GOTERM_BP_DIRECT | GO:0010862~positive regulation of pathway-restricted SMAD protein phosphorylation | 5 | 0.58 | 0.03 | 3.67 | 1 | 0.99 | 42.5 |
| GOTERM_BP_DIRECT | GO:0002062~chondrocyte differentiation | 5 | 0.58 | 0.03 | 3.67 | 1 | 0.99 | 42.5 |
| GOTERM_BP_DIRECT | GO:0046854~phosphatidylinositol phosphorylation | 9 | 1.04 | 0.04 | 2.2 | 1 | 0.99 | 49 |
| GOTERM_BP_DIRECT | GO:0045740~positive regulation of DNA replication | 7 | 0.81 | 0.04 | 2.57 | 1 | 0.99 | 50.7 |
| GOTERM_BP_DIRECT | GO:0071260~cellular response to mechanical stimulus | 10 | 1.16 | 0.04 | 2.06 | 1 | 0.99 | 51.5 |
| GOTERM_BP_DIRECT | GO:0030049~muscle filament sliding | 12 | 1.39 | 0.04 | 1.87 | 1 | 0.99 | 53.6 |
| GOTERM_BP_DIRECT | GO:0008544~epidermis development | 6 | 0.69 | 0.04 | 2.8 | 1 | 1 | 56.5 |
| GOTERM_BP_DIRECT | GO:0050999~regulation of nitric- oxide synthase activity | 6 | 0.69 | 0.04 | 2.8 | 1 | 1 | 56.5 |
| GOTERM_BP_DIRECT | GO:0016337~single organismal cell-cell adhesion | 11 | 1.27 | 0.05 | 1.89 | 1 | 1 | 61.2 |
| GOTERM_BP_DIRECT | GO:0008016~regulation of heart contraction | 5 | 0.58 | 0.05 | 3.21 | 1 | 1 | 61.3 |
| GOTERM_BP_DIRECT | GO:0048536~spleen development | 5 | 0.58 | 0.05 | 3.21 | 1 | 1 | 61.3 |
| GOTERM_BP_DIRECT | GO:0006954~inflammatory response | 27 | 3.13 | 0.05 | 1.42 | 1 | 1 | 61.4 |
| GOTERM_BP_DIRECT | GO:0030178~negative regulation of Wnt signaling pathway | 4 | 0.46 | 0.05 | 4.11 | 1 | 1 | 63.1 |
| GOTERM_BP_DIRECT | GO:0034427~nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5' | 4 | 0.46 | 0.05 | 4.11 | 1 | 1 | 63.1 |
| GOTERM_BP_DIRECT | GO:0048565~digestive tract development | 4 | 0.46 | 0.05 | 4.11 | 1 | 1 | 63.1 |
| GOTERM_BP_DIRECT | GO:0042327~positive regulation of phosphorylation | 6 | 0.69 | 0.06 | 2.57 | 1 | 1 | 70.4 |
| GOTERM_BP_DIRECT | GO:0044267~cellular protein metabolic process | 13 | 1.5 | 0.07 | 1.67 | 1 | 1 | 74.3 |
| GOTERM_BP_DIRECT | GO:0048015~phosphatidylinositol- mediated signaling | 8 | 0.93 | 0.08 | 2.06 | 1 | 1 | 76 |
| GOTERM_BP_DIRECT | GO:0007613~memory | 5 | 0.58 | 0.08 | 2.86 | 1 | 1 | 77.1 |
| GOTERM_BP_DIRECT | GO:0051781~positive regulation of cell division | 5 | 0.58 | 0.08 | 2.86 | 1 | 1 | 77.1 |
| GOTERM_BP_DIRECT | GO:0033160~positive regulation of protein import into nucleus, translocation | 5 | 0.58 | 0.08 | 2.86 | 1 | 1 | 77.1 |
| GOTERM_BP_DIRECT | GO:0006955~immune response | 26 | 3.01 | 0.08 | 1.36 | 1 | 1 | 78.8 |
| GOTERM_BP_DIRECT | GO:2001238~positive regulation of extrinsic apoptotic signaling pathway | 6 | 0.69 | 0.09 | 2.37 | 1 | 1 | 81.6 |
| GOTERM_BP_DIRECT | GO:0060348~bone development | 6 | 0.69 | 0.09 | 2.37 | 1 | 1 | 81.6 |
| GOTERM_BP_DIRECT | GO:0006941~striated muscle contraction | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0001916~positive regulation of T cell mediated cytotoxicity | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0010458~exit from mitosis | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0032270~positive regulation of cellular protein metabolic process | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0032012~regulation of ARF protein signal transduction | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0045216~cell-cell junction organization | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0097296~activation of cysteine-type endopeptidase activity involved in apoptotic | 4 | 0.46 | 0.09 | 3.43 | 1 | 1 | 82.6 |
| GOTERM_BP_DIRECT | GO:0060337~type I interferon signaling pathway | 9 | 1.04 | 0.09 | 1.85 | 1 | 1 | 83.5 |
| GOTERM_BP_DIRECT | GO:0016180~snRNA processing | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
| GOTERM_BP_DIRECT | GO:0010907~positive regulation of glucose metabolic process | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
| GOTERM_BP_DIRECT | GO:0035313~wound healing, | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
| GOTERM_BP_DIRECT | GO:0045446~endothelial cell differentiation | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
| GOTERM_BP_DIRECT | GO:0030901~midbrain development | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
| GOTERM_BP_DIRECT | GO:0048535~lymph node development | 3 | 0.35 | 0.1 | 5.14 | 1 | 1 | 84.8 |
Table 3: Gene ontology analysis showed an enrichment for protein abundance changes in osteogenesis-related biological processes, including “Extracellular matrix organization” (p = 0.001154) and “Negative regulation of TGF-beta receptor signalling” (p = 0.001894).
| Accession | Description | Gene Symbol | HU80: AHBC/native | HU82: AHBC/native | HU84: AHBC/native | Abundance Ratio Adj. P-Value: (127) / (126) | Abundance Ratio Adj. P-Value: (128) / (129) | Abundance Ratio Adj. P-Value: (131) / (130) |
|---|---|---|---|---|---|---|---|---|
| P00918 | Carbonic anhydrase 2 | CA2 | 0.95 | 1.39 | 1.34 | 0.9 | 0.8 | 0.69 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=CA2 PE=1 SV=2 | ||||||||
| Q13488 | V-type proton ATPase 116 | TCIRG1 | 1.05 | 1.07 | 0.98 | 1 | 0.96 | 1 |
| kDa subunit a isoform 3 | ||||||||
| OS=Homo sapiens OX=9606 | ||||||||
| GN=TCIRG1 PE=1 SV=3 | ||||||||
| Q9Y6K9 | NF-kappa-B essential | IKBKG | 1.15 | 0.85 | 0.99 | 0.87 | 0.96 | 0.99 |
| modulator OS=Homo | ||||||||
| sapiens OX=9606 GN=IKBKG | ||||||||
| PE=1 SV=2 | ||||||||
| A0AVT1 | Ubiquitin-like modifier- | UBA6 | 1.14 | 1.04 | 1.06 | 0.96 | 1 | 0.99 |
| activating enzyme 6 | ||||||||
| OS=Homo sapiens OX=9606 | ||||||||
| GN=UBA6 PE=1 SV=1 | ||||||||
| O75718 | Cartilage-associated protein | CRTAP | 1.07 | 0.94 | 1.01 | 1 | 1 | 0.98 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=CRTAP PE=1 SV=1 | ||||||||
| A0A1B0GUS4 | Ubiquitin-conjugating | |||||||
| enzyme E2 L5 OS=Homo | ||||||||
| sapiens OX=9606 | UBE2L5P; | |||||||
| GN=UBE2L5 PE=2 SV=1 | UBE2L5 | 1.39 | 1.62 | 1.48 | 0.27 | #### | 0.45 | |
| P02452 | OS=Homo sapiens OX=9606 | COL1A1 | 1.48 | 1.45 | 0.76 | 0.08 | 0.71 | 0.77 |
| GN=COL1A1 PE=1 SV=5 | ||||||||
| A0A075B6K4 | Immunoglobulin lambda | IGLV3-10 | 1.05 | 1.11 | 0.82 | 1 | 0.94 | 0.98 |
| variable 3-10 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=IGLV3-10 PE=3 SV=2 | ||||||||
| P08123 | Collagen alpha-2(I) chain | COL1A2 | 1.43 | 1.12 | 0.74 | 0.15 | 0.98 | 0.72 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=COL1A2 PE=1 SV=7 | ||||||||
| A0A0C4DH68 | Immunoglobulin kappa | IGKV2-24 | 1.15 | 1.65 | 1.35 | 0.95 | 0.12 | 0.5 |
| variable 2-24 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=IGKV2-24 PE=3 SV=1 | ||||||||
| Q75V66 | Anoctamin-5 OS=Homo | ANO5 | 1.26 | 1.4 | 1.14 | 0.58 | 0.6 | 0.91 |
| sapiens OX=9606 GN=ANO5 | ||||||||
| PE=1 SV=1 | ||||||||
| A0A0B4J1U7 | Immunoglobulin heavy | IGHV6-1 | 0.97 | 0.71 | 0.5 | 0.96 | 0.79 | 0.04 |
| variable 6-1 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=IGHV6-1 PE=3 SV=1 | ||||||||
| P07237 | Protein disulfide-isomerase | P4HB | 1 | 1.24 | 1.21 | 0.97 | 0.94 | 0.89 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=P4HB PE=1 SV=3 | ||||||||
| A0A0B4J2D5 | Glutamine | |||||||
| amidotransferase-like class | ||||||||
| 1 domain-containing protein | ||||||||
| 3B, mitochondrial OS=Homo | ||||||||
| sapiens OX=9606 | C21orf33; | |||||||
| GN=GATD3B PE=1 SV=1 | LOC102724023 | 1.02 | 1.11 | 1.11 | 0.99 | #### | 0.97 | |
| P36955 | Pigment epithelium-derived | SERPINF1 | 1.1 | 1.16 | 0.85 | 0.99 | 0.96 | 0.95 |
| factor OS=Homo sapiens | ||||||||
| OX=9606 GN=SERPINF1 | ||||||||
| PE=1 SV=4 | ||||||||
| A1L4H1 | Soluble scavenger receptor | SSC5D | 1.08 | 0.7 | 0.57 | 0.98 | 0.95 | 0.69 |
| cysteine-rich domain- | ||||||||
| containing protein SSC5D | ||||||||
| OS=Homo sapiens OX=9606 | ||||||||
| GN=SSC5D PE=1 SV=3 | ||||||||
| O00469 | Procollagen-lysine,2- | PLOD2 | 1.18 | 0.76 | 0.95 | 0.85 | 0.98 | 0.98 |
| oxoglutarate 5-dioxygenase | ||||||||
| 2 OS=Homo sapiens | ||||||||
| OX=9606 GN=PLOD2 PE=1 | ||||||||
| SV=2 | ||||||||
| A0A075B767 | Peptidyl-prolyl cis-trans | LOC101060723; | ||||||
| LOC100996754; | ||||||||
| isomerase A-like 4H | LOC105371242 | 1.35 | 1.68 | 1.29 | 0.43 | #### | 0.62 | |
| P23284 | OS=Homo sapiens OX=9606 | PPIB | 1.13 | 1.24 | 1.17 | 0.97 | 0.94 | 0.93 |
| GN=PPIAL4H PE=3 SV=1 | ||||||||
| Peptidyl-prolyl cis-trans | ||||||||
| isomerase B OS=Homo | ||||||||
| sapiens OX=9606 GN=PPIB | ||||||||
| PE=1 SV=2 | ||||||||
| A0FGR8 | Extended synaptotagmin-2 | ESYT2 | 0.98 | 1.04 | 0.99 | 0.99 | 0.97 | 0.99 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=ESYT2 PE=1 SV=1 | ||||||||
| Q96AY3 | Peptidyl-prolyl cis-trans | FKBP10 | 1.01 | 1.04 | 0.83 | 0.99 | 0.98 | 0.95 |
| isomerase FKBP10 | ||||||||
| OS=Homo sapiens OX=9606 | ||||||||
| GN=FKBP10 PE=1 SV=1 | ||||||||
| A0A087WW87 | Immunoglobulin kappa | IGKV2-40; | ||||||
| variable 2-40 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| Q32P28 | Prolyl 3-hydroxylase 1 | LEPRE1; P3H1 | 1.15 | 0.59 | 0.71 | 0.93 | 0.74 | 0.9 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=P3H1 PE=1 SV=2 | ||||||||
| A1L0T0 | Acetolactate synthase-like | ILVBL | 1.05 | 0.85 | 0.88 | 1 | 0.97 | 0.99 |
| protein OS=Homo sapiens | ||||||||
| OX=9606 GN=ILVBL PE=1 | ||||||||
| SV=2 | ||||||||
| P09486 | SPARC OS=Homo sapiens | SPARC | 1.51 | 0.97 | 0.89 | 0.05 | 0.97 | |
| OX=9606 GN=SPARC PE=1 | ||||||||
| SV=1 | 0.97 | |||||||
| A0MZ66 | Shootin-1 OS=Homo sapiens | KIAA1598; | ||||||
| OX=9606 GN=SHTN1 PE=1 | ||||||||
| SV=4 | SHTN1 | 1.24 | 1.38 | 1.36 | 0.66 | #### | 0.64 | |
| Q9UQ90 | Paraplegin OS=Homo | SPG7 | 1.08 | 1.04 | 0.83 | 0.99 | 0.95 | 0.99 |
| sapiens OX=9606 GN=SPG7 | ||||||||
| PE=1 SV=2 | ||||||||
| A0A0B4J2D9 | Immunoglobulin kappa | IGKV1D-13; | ||||||
| variable 1D-13 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=IGKV1D-13 PE=3 SV=1 | IGKV1-13 | 0.88 | 0.55 | 0.85 | 0.78 | #### | 0.99 | |
| P50454 | Serpin H1 OS=Homo sapiens | SERPINH1 | 1.14 | 0.95 | 0.79 | 0.96 | 0.96 | 0.82 |
| OX=9606 GN=SERPINH1 | ||||||||
| PE=1 SV=2 | ||||||||
| A0A0C4DH38 | Immunoglobulin heavy | IGHV5-51 | 1 | 1.18 | 0.89 | 0.99 | 0.88 | 0.99 |
| variable 5-51 OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=IGHV5-51 PE=3 SV=1 | ||||||||
| O94855 | Protein transport protein | SEC24D | 1.02 | 0.85 | 0.81 | 1 | 0.97 | 0.96 |
| Sec24D OS=Homo sapiens | ||||||||
| OX=9606 GN=SEC24D PE=1 | ||||||||
| SV=2 | ||||||||
| Q16610 | Extracellular matrix protein | ECM1 | 1.03 | 0.96 | 0.72 | 1 | 1 | 0.85 |
| 1 OS=Homo sapiens | ||||||||
| OX=9606 GN=ECM1 PE=1 | ||||||||
| SV=2 | ||||||||
| Q4V9L6 | Transmembrane protein | TMEM119 | 1.71 | 0.99 | 0.95 | 0 | 0.96 | 0.99 |
| 119 OS=Homo sapiens | ||||||||
| OX=9606 GN=TMEM119 | ||||||||
| PE=1 SV=1 | ||||||||
| P01344 | Insulin-like growth factor II | IGF2 | 1.73 | 1.23 | 1.09 | 0 | 0.83 | 0.95 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=IGF2 PE=1 SV=1 | ||||||||
| P02788 | Lactotransferrin OS=Homo | LTF | 1.12 | 0.98 | 1.13 | 0.98 | 0.98 | 0.97 |
| sapiens OX=9606 GN=LTF | ||||||||
| PE=1 SV=6 | ||||||||
| P08493 | Matrix Gla protein | MGP | 1.5 | 0.69 | 0.87 | 0.17 | 0.95 | 0.99 |
| OS=Homo sapiens OX=9606 | ||||||||
| GN=MGP PE=1 SV=2 | ||||||||
| Q99523 | Sortilin OS=Homo sapiens | SORT1 | 0.8 | 0.8 | 0.84 | 0.33 | 0.97 | 0.99 |
| OX=9606 GN=SORT1 PE=1 | ||||||||
| SV=3 | ||||||||
| Q9Y240 | C-type lectin domain family | CLEC11A | 1.32 | 0.97 | 0.75 | 0.37 | 1 | 0.78 |
| 11 member A OS=Homo | ||||||||
| sapiens OX=9606 | ||||||||
| GN=CLEC11A PE=1 SV=1 | ||||||||
| P34820 | Bone morphogenetic | BMP8B | 1.24 | 0.48 | 0.67 | 0.68 | 0.35 | 0.87 |
| protein 8B OS=Homo | ||||||||
| Q9NR12 | sapiens OX=9606 | PDLIM7 | 1.16 | 0.51 | 0.92 | 0.9 | 0.01 | 0.99 |
Table 4: Focusing on bone-related proteins as annotated by the UniProt knowledgebase specific classes of proteins appeared to exhibit concordant abundance changes across the 3 biological replicates.
Figure 3: (A) Proteomic profiling of human AHBC shows similarity in proteomic profiles between AHBC and donor-matched native bone specimens (n=3). The 865 proteins exhibiting ≥1.5-fold difference (up or down) between AHBC and donor-matched native bone in at least 1 donor were used as inputs for hierarchical clustering. Protein abundance was color-coded based on Log10-transformed peptide ion intensity values, summed up to the individual parent proteins. (B) Relative abundance changes of bone-related proteins in human AHBC vs. donor-matched native bone specimens. 40 proteins with bone-related UniProt keywords were used as inputs for hierarchical clustering (Table S4). Relative protein abundance changes are represented and color-coded as fold-changes. Principal component analysis of transcriptomic profiles from (C) 3 (angiogenesis) and (D) 4 (wound healing) technical replicates.
Proteins exhibiting abundance increases post-processing were enriched for gene ontology terms including ECM organization, endochondral ossification, and osteoblast differentiation (Figures 4b, 5A), indicating possible contributions to bone regeneration at the proteomic level. In line with the proteomic profiles, transcriptomic profiles of cranial bone assessed immediately after processing also demonstrated clear separation between native and processed samples (Figures 5B, 5C, 5D). PCA of wound healing, angiogenesis, and osteogenesis pathway transcripts in the post- processed AHBC appeared to be distinct from the matching pre-processing samples suggesting a priming effect of AHBC processing (Figure 5D).
Figure 5: (A) Heatmap for proteomic abundance profiles of AHBC and donor-matched rabbit cranial bone (n=4). Dark purple: highest abundance; white: lowest. AHBC from 3 out of 4 rabbits clustered away from native bone specimens. Principal component analysis (PCA) of transcripts associated with (B) wound healing, (C) angiogenesis, (D) osteogenesis pathways. The first and second principal components (PCs) are shown on the x and y axis, respectively.
Evaluation of AHBC Treatment in Pre-Clinical Models
All animals survived the 8-week study duration without adverse clinical signs or experimental complications. Antemortem evaluations conducted at least twice daily over the duration of each study reported appropriate post-operative recovery times, effective pain management (analgesia) and supportive care, stable body condition with progressive weight gain, well tolerated general handling and physical examinations. There were no neurologic and musculoskeletal reports to suggest complications associated model induction or intervention procedures. Post-mortem evaluations were performed by a qualified veterinarian blinded to experimental group assignments. There was no evidence of infection, hematoma, or necrosis at or around cranial defect or lumbar spinal fusion sites.
Specific to the cranial defect study, the dura mater and brain tissue under lying each defect site was free from evidence of acute or chronic inflammation, adhesions, scar formation, or adverse tissue reactions associated with bone grafting materials.
Specific to the posterolateral lumbar fusion study, there was no evidence of damage to the paraspinal musculature, adjacent lumbar processes or vertebral bodies.
Rabbit AHBC-regenerated cranial bone has similar architecture and bone mineral density as native bone
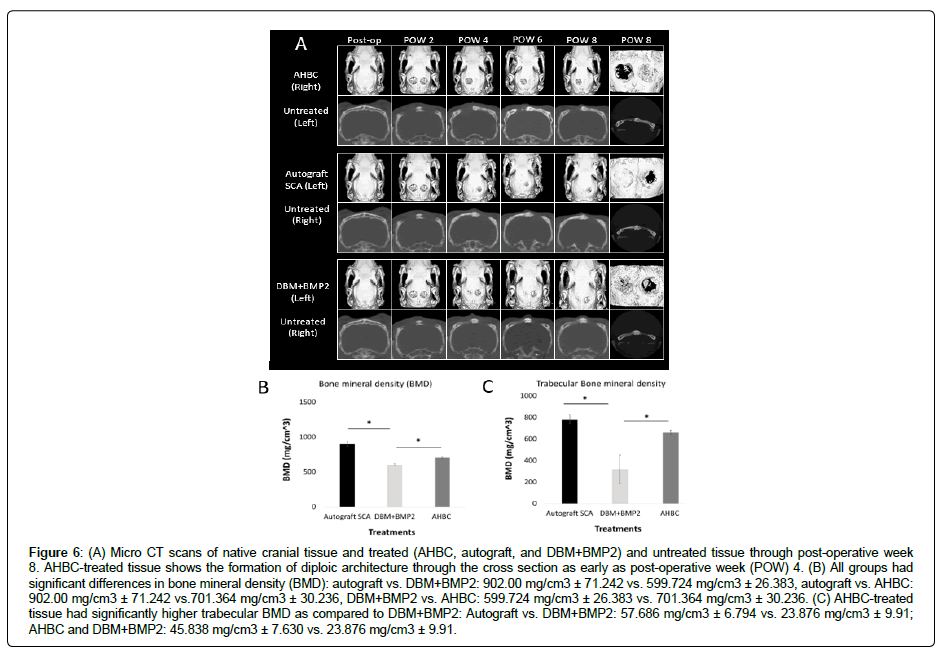
Serial μCT imaging revealed closure of cranial defects beginning at post-operative week 4 (POW4) in AHBC-treated, SCA, and DBM+BMP2 groups. Closure of defects was not seen in untreated internal control groups (Figure 6A). AHBC-treated tissue displayed diploic bone architecture at POW4, while SCA, and DBM+BMP2 groups did not show diploic bone formation. Bone mineral density (BMD) (Figure 6B) was measured between groups using a oneway ANOVA with p < 0.001. Significant differences were found between all groups using a Tukey’s multiple comparison test: SCA vs. DBM+BMP2: 902.00 mg/cm3 ± 71.242 vs. 599.724 mg/cm3 ± 26.383, p < 0.001; SCA vs. AHBC: 902.00 mg/cm3 ± 71.242 vs.701.364 mg/ cm3 ± 30.236, p < 0.001; DBM+BMP2 vs. AHBC: 599.724 mg/cm3 ± 26.383 vs. 701.364 mg/cm3 ± 30.236, p = 0.0134. Trabecular bone mineral density was also assessed (Figure 6C) using an ANOVA oneway analysis with p = 0.0020. No significant difference was observed between SCA and AHBC (57.686 mg/cm3 ± 6.794 vs. 45.838 mg/ cm3 ± 7.630, p = 0.4960). However, significant differences were seen between SCA and DBM+BMP2 (57.686 mg/cm3 ± 6.794 vs. 23.876 mg/cm3 ± 9.91, p = 0.0020) and between AHBC and DBM+BMP2 (45.838 mg/cm3 ± 7.630 vs. 23.876 mg/cm3 ± 9.91, p = 0.0155)
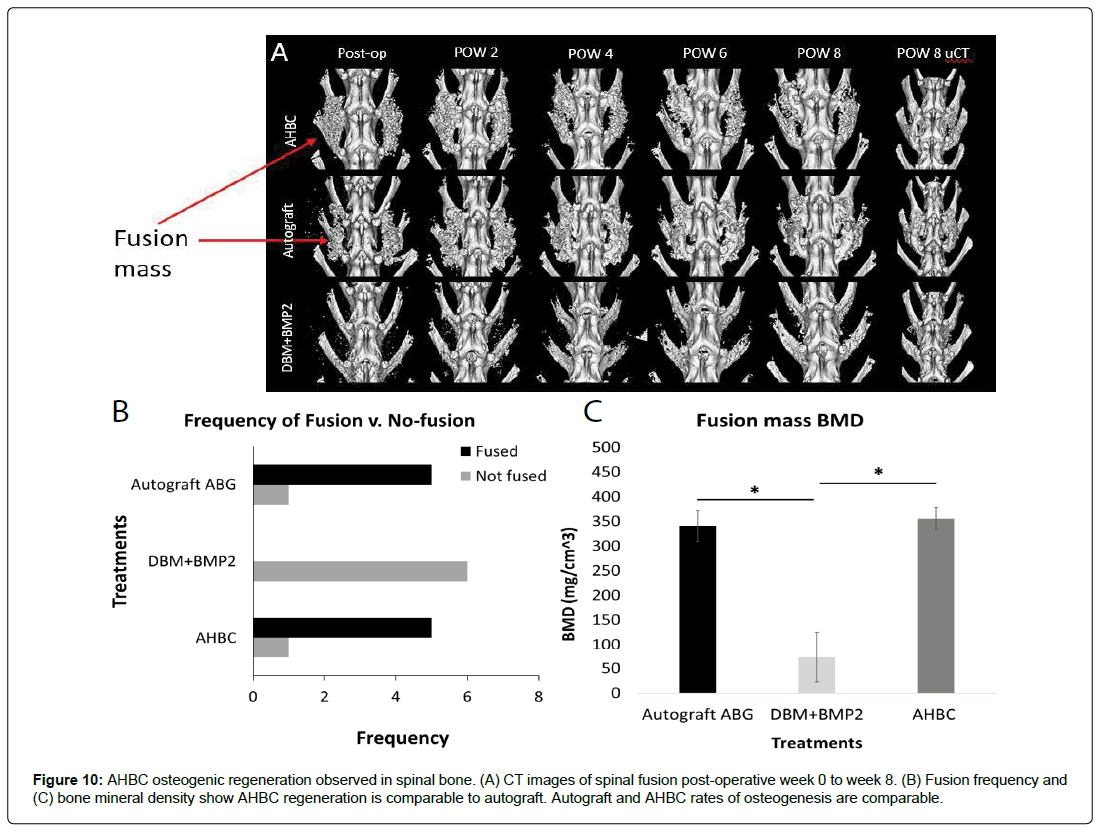
Figure 6: (A) Micro CT scans of native cranial tissue and treated (AHBC, autograft, and DBM+BMP2) and untreated tissue through post-operative week 8. AHBC-treated tissue shows the formation of diploic architecture through the cross section as early as post-operative week (POW) 4. (B) All groups had significant differences in bone mineral density (BMD): autograft vs. DBM+BMP2: 902.00 mg/cm3 ± 71.242 vs. 599.724 mg/cm3 ± 26.383, autograft vs. AHBC: 902.00 mg/cm3 ± 71.242 vs.701.364 mg/cm3 ± 30.236, DBM+BMP2 vs. AHBC: 599.724 mg/cm3 ± 26.383 vs. 701.364 mg/cm3 ± 30.236. (C) AHBC-treated tissue had significantly higher trabecular BMD as compared to DBM+BMP2: Autograft vs. DBM+BMP2: 57.686 mg/cm3 ± 6.794 vs. 23.876 mg/cm3 ± 9.91; AHBC and DBM+BMP2: 45.838 mg/cm3 ± 7.630 vs. 23.876 mg/cm3 ± 9.91.
Rabbit AHBC-treated and SCA treated cranial defects have significantly different mechanical properties compared to DBM+BMP2 treated defects
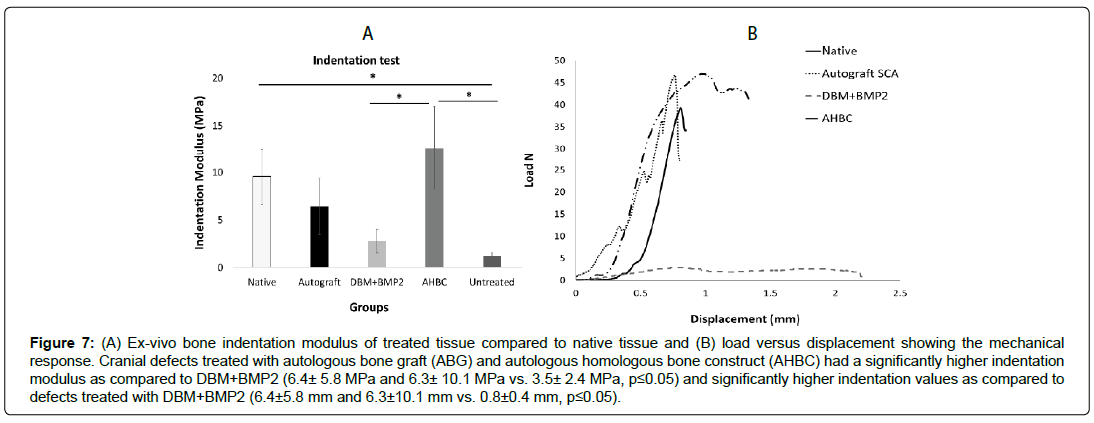
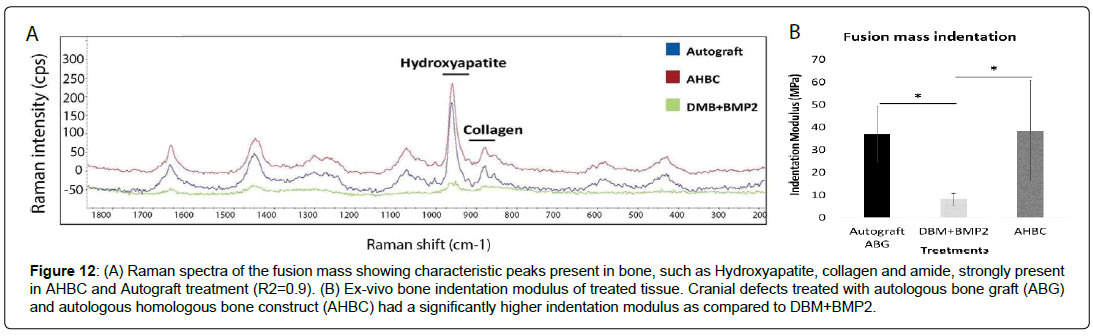
Ex-vivo bone indentation modulus and load versus displacement testing of AHBC-treated tissue was compared to DBM+BMP2, SCA, and native tissue groups using a universal testing machine (Figure 7A, B). Cranial defects treated with SCA and AHBC had a significantly higher indentation modulus as compared to DBM+BMP2 (6.4 ± 5.8 MPa and 6.3 ± 10.1 MPa vs. 3.5 ± 2.4 MPa, p ≤ 0.05). Native tissue, AHBC-treated tissue, and SCA treated groups were comparable (no significant differences between groups) in stiffness measurements. DBM+BMP2 had significantly lower mechanical measurements than all other groups.
Figure 7: (A) Ex-vivo bone indentation modulus of treated tissue compared to native tissue and (B) load versus displacement showing the mechanical response. Cranial defects treated with autologous bone graft (ABG) and autologous homologous bone construct (AHBC) had a significantly higher indentation modulus as compared to DBM+BMP2 (6.4± 5.8 MPa and 6.3± 10.1 MPa vs. 3.5± 2.4 MPa, p≤0.05) and significantly higher indentation values as compared to defects treated with DBM+BMP2 (6.4±5.8 mm and 6.3±10.1 mm vs. 0.8±0.4 mm, p≤0.05).
Rabbit AHBC-treated cranial bone has similar hydroxyapatite levels as native and split calvarial autograft tissue
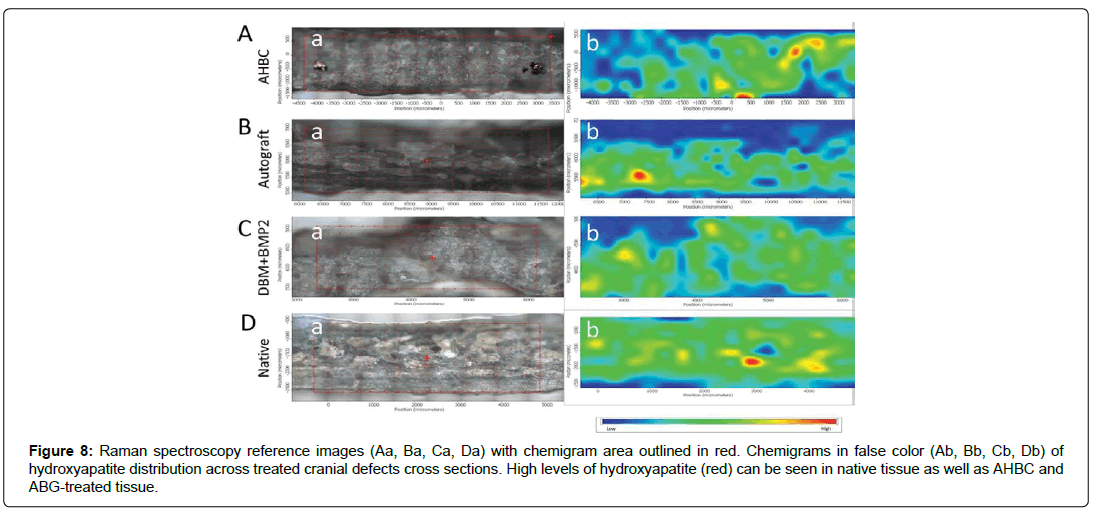
Bone mineral hydroxyapatite levels were obtained using Raman spectroscopy on ex-vivo cranial cross sections (Figure 8). Raman spectroscopy reference areas (Figures 8Aa, 8Ba, 8Ca, 8Da) were obtained for each treatment group; red boxes show areas utilized for chemigrams (Figures 8Ab, 8Bb, 8Cb, 8Db). Chemigrams are shown in false color and depict distribution of hydroxyapatite, from low levels (blue) to high levels (red). AHBC-treated cranial bone displayed high amounts of hydroxyapatite (Figure 8Ab) comparable to amounts seen in SCA treated defects (Figure 8Bb) and native tissue (Figure 8Db). DBM+BMP2 had relatively low levels of hydroxyapatite (Figure 8Cb).
Figure 8: Raman spectroscopy reference images (Aa, Ba, Ca, Da) with chemigram area outlined in red. Chemigrams in false color (Ab, Bb, Cb, Db) of hydroxyapatite distribution across treated cranial defects cross sections. High levels of hydroxyapatite (red) can be seen in native tissue as well as AHBC and ABG-treated tissue.
Rabbit AHBC-treated cranial bone displays ultrastructure similar to native cranial bone
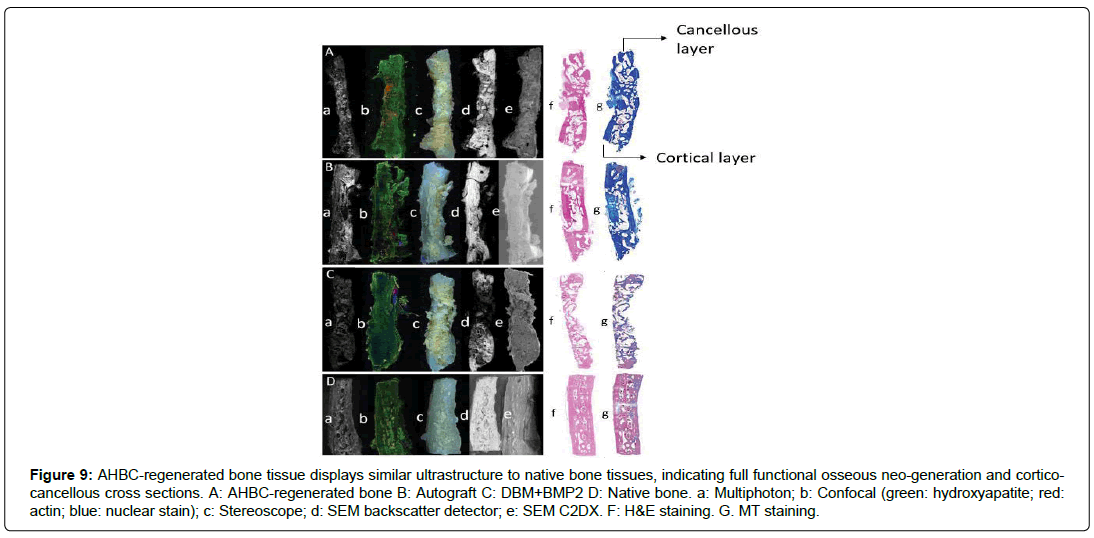
Multiple imaging techniques were utilized to evaluate the ultrastructure of all treatment groups and native cranial tissue ex-vivo, including multiphoton imaging, confocal imaging, stereoscope, SEM with backscatter detection, and SEM C2DX modalities. Multiphoton imaging shows cortical and cancellous bone and diploic architecture in AHBC-treated cranial bone (Figure 9Aa), comparable to that of native bone (Figure 9Da), and absent in SCA (Figure 9Ba) and DBM+BMP2 bone (Figure 9Ca). Collagen presence and arrangement can also be observed on multiphoton imaging, with SCA (Figure 9Ba) and AHBC-treated bone (Figure 9Aa) similar to native bone (Figure 9Da). Collagen levels of DBM+BMP2 (Figure 9Ca) are lower than those of other groups. Confocal microscopy was performed to evaluate mineralization and hydroxyapatite deposition by osteogenic cells, actin, and nuclei (Figures 9Ab-Db). Hydroxyapatite appears green, actin appears red, and nuclei appear blue as observed with confocal microscopy. Hydroxyapatite is prominent in AHBC-treated bone (Figure 9Ab), SCA treated tissue (Figure 9Bb), and native cranial bone (Figure 9Db) and less apparent in DBM+BMP2 (Figure 9Cb). Actin is much more pronounced in AHBC cranial bone and native tissue (Figures 9Ab, 9Db) than in all other groups (Figures. 9Bb, 9Cb).
Figure 9: AHBC-regenerated bone tissue displays similar ultrastructure to native bone tissues, indicating full functional osseous neo-generation and corticocancellous cross sections. A: AHBC-regenerated bone B: Autograft C: DBM+BMP2 D: Native bone. a: Multiphoton; b: Confocal (green: hydroxyapatite; red: actin; blue: nuclear stain); c: Stereoscope; d: SEM backscatter detector; e: SEM C2DX. F: H&E staining. G. MT staining.
Stereoscopic imaging revealed AHBC (Figure 9Ac) and DBM+BMP2 treated defects (Figure 9Cc) appear to have dense neoosteogenic deposits as compared to SCA treatment (Figure 9Dc).
However, SEM imaging with backscatter detection shows DBM+BMP2 (Figure 9Cd) to be the least dense bone of all groups, with SCA (Figure 9Ad) and AHBC (Figure 9Ad) appearing similar to the density of native cranial tissue (Figure 9Dd). SEM C2DX shows all neo-generated tissue groups (Figures 9Ae, 9Be, and 9Ce) have distinct architecture from native cranial tissue (Figure 9De). While SCA appears most similar to native tissue in texture, it lacks the diploic architecture seen in native tissue and observed in CT scans of AHBC-treated tissue (Figure 5a).
Rabbit AHBC osteogenesis observed in spinal bone is similar to that of autologous bone graft (ABG)
Spinal fusion was observed in 8-weeks post-operative CT scans. Defect areas were treated with AHBC, ABG, or DBM+BMP2 and subsequently imaged (Figure 10A) shows scans POW 0-8). There was a significant difference in the size of spinal fusion masses between AHBC treatment and DBM+BMP2 treated defects as observed during bi-weekly CT scans (Figure 10A). ABG-treated fusion masses and AHBC treated fusion masses had an average palpation score per treatment of 1.83 (n=6) while DBM+BMP2 treated fusion masses had an average score of 0.33 (n=6). The frequency of spinal fusion in each group was also noted (Figure 10B) with both ABG and AHBC treatment groups having 5 fusions and 1 non-fusion (n=6); and DBM+BMP2 being the least effective with 0 fusion, 2 partial fusions and 4 non-fusions (n=6).
Ex-vivo bone mineral density was obtained from treated defects and quantitated (Figure 10C). One-way ANOVA analysis demonstrated significant differences between groups (p < 0.0001). Bone mineral density measurements were compared to ABG using a Tukey’s multiple comparison test. AHBC and ABG were not significantly different, while both were significantly different from DBM+BMP2: ABG vs. AHBC: 340.3308 mg/cm3 ± 62.54099 vs. 355.4813 mg/cm3 ± 44.77283, p = 0.8715; ABG vs. DBM+BMP2: 340.3308 mg/cm3 ± 62.54099 vs 73.3683 mg/cm3 ± 102.8192, p <0.001; AHBC vs. DBM+BMP2: 355.4813 mg/cm3 ± 44.77283 vs. 73.3683 mg/cm3 ± 102.8192, p <0.001. This trend is noticeable in μCT scans of all groups at 8 weeks post-operative (Figure 10A), as AHBC and ABG treatment show bigger and denser fusion masses than DBM+BMP2 treatment (Figure 10C).
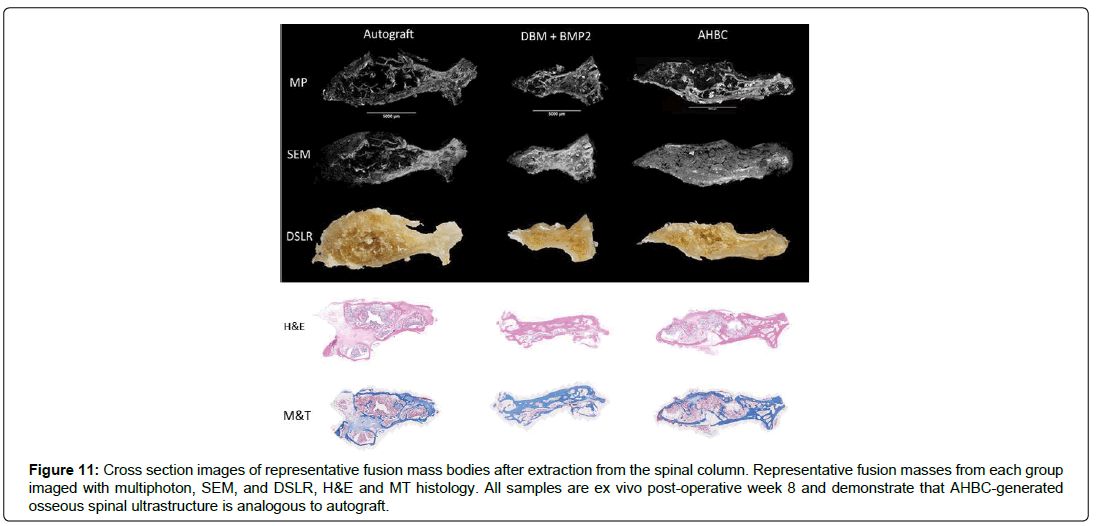
Rabbit AHBC treated spinal fusion shows ultrastructure similarities to Rabbit ABG generated spinal bone
To observe osteogenesis and ultrastructure of AHBC, ABG, and DBM+BMP2, multiple imaging modalities were employed on excised samples. Representative samples from each treatment group were imaged at POW 8. Inter-vertebral sections (Figure 11) display large areas of fusion in AHBC and ABG groups. Cortical and cancellous bone architecture appears analogous between AHBC and ABG groups, with DBM+BMP2 showing minimal osteogenesis and fibrous appearance as observed based on Raman spectra of the fusion mass (Figure 12). Ex-vivo cross sections of fusion mass bodies of vertebrae (Figure 11) show the differences between DBM+BMP2 and the ABG and AHBC treatment groups.
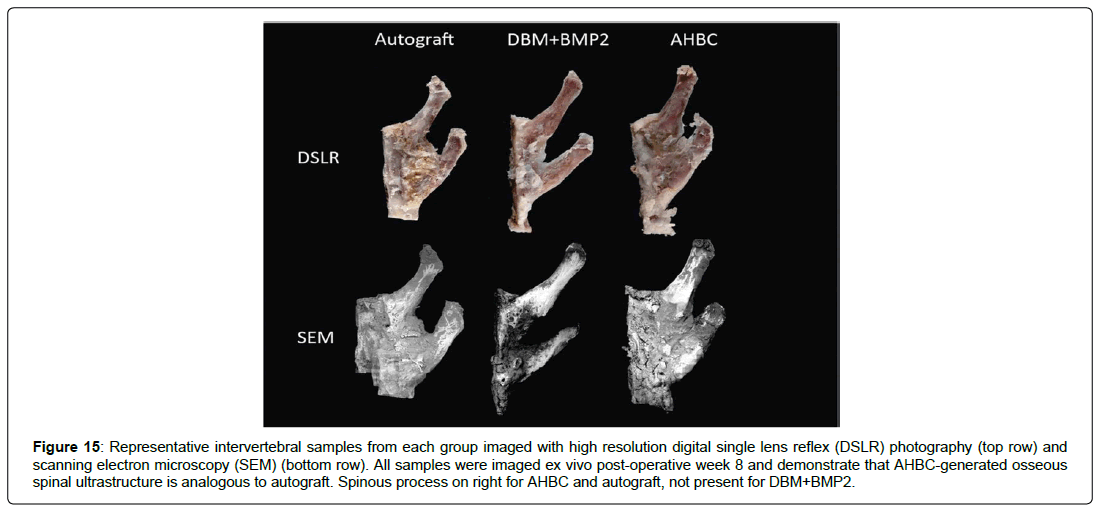
Figure 11: Cross section images of representative fusion mass bodies after extraction from the spinal column. Representative fusion masses from each group imaged with multiphoton, SEM, and DSLR, H&E and MT histology. All samples are ex vivo post-operative week 8 and demonstrate that AHBC-generated osseous spinal ultrastructure is analogous to autograft.
Figure 12: (A) Raman spectra of the fusion mass showing characteristic peaks present in bone, such as Hydroxyapatite, collagen and amide, strongly present in AHBC and Autograft treatment (R2=0.9). (B) Ex-vivo bone indentation modulus of treated tissue. Cranial defects treated with autologous bone graft (ABG) and autologous homologous bone construct (AHBC) had a significantly higher indentation modulus as compared to DBM+BMP2.
The molecular profile of Rabbit AHBC-generated bone is similar to that of Rabbit ABG- generated bone or native bone
We assessed osteogenesis, wound healing, angiogenesis, and extra cellular matrix pathways of the bone resulting from AHBC and ABG treatment as well as native bone 8 weeks post-study initiation. Comparisons of the molecular profiles for each of the 4 pathways demonstrated that AHBC-generated fusion mass had a similar expression profile from was paired native or ABG- generated fusion samples at 8 weeks post treatment (data not shown), consistent with the physiological data. Paired non-parametric t-tests comparing native to AHBC or native to ABG did not yield statistically significant associations (PBonferroni < 5.88x10-4; data not shown).
These data suggest that AHBC-treatment results in bone tissues with native expression profiles.
Discussion
The goal of this study was to evaluate the effect of AHBC processing has on human and animal tissue and whether it can generate bone in large animal models. This study demonstrated that (a) AHBC processing primes the tissue by altering osteogenicrelated gene transcription and protein profiles and retains relevant structural characteristics of bone; (b) AHBC can regenerate diploic cortico-cancellous bone in cranial defect and spinal fusion models; (c) the regenerated defects have similar bone mineral density and mechanical strength as native bone; (d) AHBC generated tissues show similar ultrastructure with consistent presence of hydroxyapatite and collagen comparable to native bone; and (e) healed AHBC treated defects achieve a similar expression profile (osteogenesis, wound healing) as autograft treated defects and native bone. Overall, functional outcomes for cranioplasty [30] and spinal arthrodesis model were chosen based on ASTM guidelines and previously published literature [24,31].
Autograft has been the enduring gold standard for cranioplasty [32] and spinal fusion [33,34], though concerns of donor-site size limitations and complications [35] lead to the use of allograft bone, alloplastic material, and bone substitutes. A wide variety of allogenic materials have been developed; however, they have higher rates of infection6, graft failure [36], graft resorption [37], or failure of complete fusion [38]. Bone substitutes have been developed and evolved in response to the shortcomings of autografts, and allografts. They are classified into ceramic-based [39], polymer- based [40,41], or growth factor-based [16,42] bone grafts. Ceramic and polymer-based products are relatively inexpensive, uniform in quality, unlimited in supply, and lack the risk of disease transmission. However, they are prone to infection and require removal when it occurs. The most widely used polymer-based product is polymethyl methacrylate (PMMA) [43]. Typically, it is used as a bone cement and filler in orthopaedic and craniofacial applications. It functions to increase load-bearing of reconstructed areas. As with other ceramic-based and polymer-based products, PMMA is very dense with low porosity thus preventing bone ingrowth.
To address the disadvantages of ceramic and polymer-based bone substitutes, tissue engineering techniques have been applied to develop several growth factor-based products. Currently, only two commercially produced growth factors have been FDA approved for clinical use: bone morphogenetic protein-2 (BMP-2) and bone morphogenetic protein-7 (BMP-7) [44,45]. They are not used alone but combined with a structural component such as DBM or collagen scaffolds, and they act primarily as cytokines mediating the differentiation of mesenchymal cells into cartilage and bone forming cells. Use of BMPs, however, have been associated with increased inflammation [46], swelling [47], and concern for tumorigenesis [48].
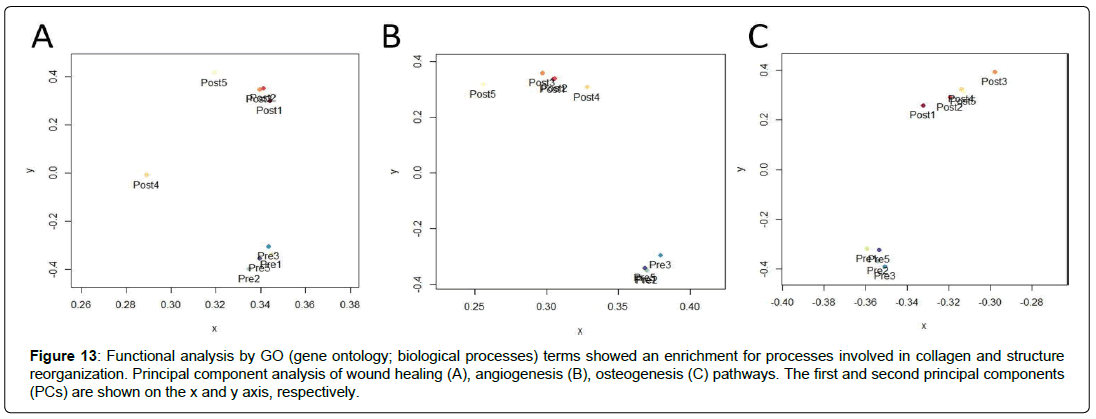
An AHBC has been developed to address bone defects. It is sourced from a small piece of autologous healthy bone. The preparation of AHBC seeks to minimize donor site morbidity and maximize de novo bone growth. Additionally, the AHBC prepared for this study from human and rabbit sources demonstrated transcriptomic and proteomic changes in pathways associated with osteogenesis (Figure 13). These include protein abundance increases in extracellular matrix proteins 2 (ECM2) and 13 other proteins involved in ECM organization (Figure 7A), which are associated with ECM organization and stem cell priming [49–51]. In this paper, In-vivo bone regeneration using Rabbit AHBC was evaluated in rabbit cranioplasty and spinal fusion models. Treatment effects were assessed by functional, molecular, structural, and histological outcomes such as osteogenic gene expression, mechanical strength, bone mineral density, hydroxyapatite distribution and mineral and collagen architecture. μCT imaging of Rabbit AHBC treated cranial defects showed osteogenesis exceeding DBM+BMP2 allograft, and bone ultrastructure (including cortical and cancellous bone and diploic architecture) resembling native tissue (Figure 5A). AHBCtreated cranial trabecular bone mineral density (TBMD) was similar to that of SCA treated bone and native bone (Figure 5C), and significantly different from DBM+BMP2. SEM C2DX imaging (Figures 6Ae & 6De) bolstered ultrastructure findings seen on μCT with AHBC-treated bone and native tissue appearing comparable in cancellous and cortical bone components, which was not seen in split-calvarial autograft, or DBM+BMP2 groups (Figures. 6Be & 6Ce) in the cranioplasty model. AHBC treated defect histomorphology had native-like cortico-cancellous diploic structure and no excessive fibrotic tissue content, which can happen with ceramic bone substitutes. A similar pattern was seen in the spinal fusion study, with AHBC treated fusion mass significantly exceeding the bone mineral density of DBM+BMP2 treated fusion mass. AHBC and ABG treatment had comparable bone mineral density which correlated with their palpation scores and the ultrastructure findings seen on SEM C2DX imaging. AHBC and ABG treated fusion mass showed presence of comparable cortical and cancellous components while DBM+BMP2 treatment had excessive fibrotic tissues and lower palpation scores (Figure 10). From a physiological perspective, the indentation modulus measurements of AHBC treated cranial defects (Figures 7A & 7B) and AHBC treated spinal fusion masses (Figure 14) were significantly higher compared to DBM+BMP-2 treatment. The mechanical strength of AHBC, SCA and ABG treated cranial defects and spinal fusion mass was similar to native bone. Matching the mechanical properties of the treated defects to the native graft environment was critically important so that progression of tissue healing is not limited by mechanical failure of the defect prior to successful tissue regeneration. The mineral component of bone, hydroxyapatite, was observed in high concentrations in AHBC, SCA, ABG, and native tissue in both Raman spectroscopy chemigrams (Figure 5) and staining viewed via confocal microscopy (Figure 6). Presence of vasculature and organized collagen architecture was seen using confocal microscopy as stained with Actin and multiphoton microscopy. Overall, structural, mechanical and anatomical outcomes obtained through AHBC application in this study were correlative and significantly better than DBM based products. These outcomes are rare for such defects treated with ceramic or synthetic bone substitutes.
Figure 13: Functional analysis by GO (gene ontology; biological processes) terms showed an enrichment for processes involved in collagen and structure reorganization. Principal component analysis of wound healing (A), angiogenesis (B), osteogenesis (C) pathways. The first and second principal components (PCs) are shown on the x and y axis, respectively.
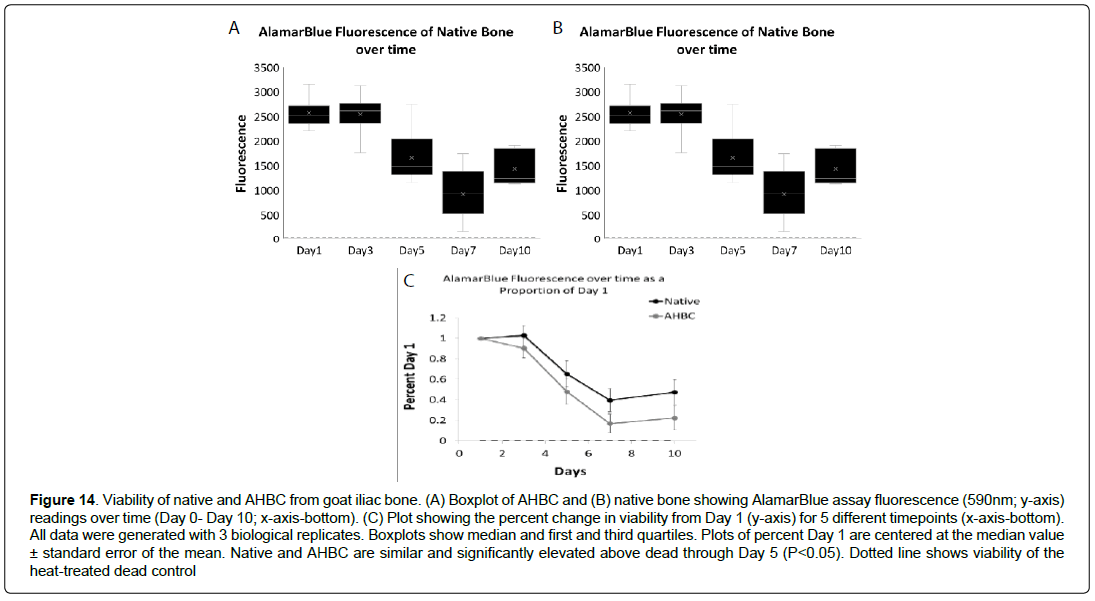
Figure 14: Viability of native and AHBC from goat iliac bone. (A) Boxplot of AHBC and (B) native bone showing AlamarBlue assay fluorescence (590nm; y-axis) readings over time (Day 0- Day 10; x-axis-bottom). (C) Plot showing the percent change in viability from Day 1 (y-axis) for 5 different timepoints (x-axis-bottom). All data were generated with 3 biological replicates. Boxplots show median and first and third quartiles. Plots of percent Day 1 are centered at the median value ± standard error of the mean. Native and AHBC are similar and significantly elevated above dead through Day 5 (P<0.05). Dotted line shows viability of the heat-treated dead control
Additionally, further studies are underway to look at effects of size and harvest site variation on AHBC functionality. These studies will explore expansion capabilities of AHBC and examine if smaller AHBC quantity could be utilized for treatment of larger defects
In conclusion, these data demonstrate that AHBC is an easily deployable graft that was able to regenerate tissue comparable in structure and function to native bone in critical-sized defect cranioplasty and spinal arthrodesis models. This translational study shows potential for AHBC as an alternative to current bone graft techniques and warrants further investigation of AHBC expansion in larger animal models.
Supplemental material
Methods AFM
Topographical and stiffness information of bone was obtained using a WITec Alpha 300RA atomic force microscope (AFM) in Digital Pulsed Force Mode (DPFM). The following settings were used to obtain native bone and AHBC topography and stiffness images: 2.8 N/m cantilever, 5 um x 5 um scan area containing 256 x 256 points, scan speed 1 s/line, retrace speed 1 s/line, maximum force setpoint 0.8 V, P-gain 3%, I-gain 5%, driving amplitude between 3-6 V, driving frequency 1 kHz, and sampling rate 1 MHz.
Viability Assay
AHBC prepared using goat iliac bone tissue was evaluated on Days 0, 1, 3, 5, 7, and 10. The number of technical replicates and timepoints were adjusted in human groups based on tissue availability. Dead controls were generated using a well-established method of autoclaving bone samples at 135°C for 20 minutes. Dead controls were used to determine the baseline of the AlamarBlue assay readouts for non-metabolically active tissue. Bone-only controls were tested by submerging bone samples in media without AlamarBlue; this tested for auto-fluorescence of the bone. AlamarBlue only blanks were obtained to provide a measurement of media contribution to fluorescence values and were subtracted from final readings. AlamarBlue assay was performed on a sample of each tissue on Day 0. Samples were incubated at 37°C in the dark for 2 hours with 10% Alamar blue. For each fluorescence reading, samples were gently vortexed and spun at 1000 G for ten seconds to clear the solution of auto-fluorescent bone particulates.100 uL media from each sample was transferred to a black-walled clear bottom 96-well plate (catalog # 07-200-567, Thermo Fisher Scientific, Waltham, MA) 100 uL of tissue-free AlamarBlue was transferred to three wells in the same black-walled clear bottom 96-well plate to serve as blank controls. Measurements were taken of the fluorescence of each sample in the plate using a plate reader (Varioskan™ multimode microplate reader, Thermo Fisher Scientific, Waltham, MA) with an excitation of 560 nm and an emission of 590 nm. All remaining timepoint samples for human and goat tissue were weighed and placed into 1.7 mL microcentrifuge tubes and performed the same way.
Blank controls were subtracted from each reading to account for the background fluorescence of the incubation media. Human and goat tissue based AHBC was assessed for differences using a 2-way ANOVA and Sidak’s multiple comparison test with a threshold of p < 0.05. Viability tissue samples were compared to dead readings using a 2-way ANOVA.
General Handling, Anaesthesia, Analgesia and Peri-Operative Monitoring
On the day of surgery rabbits were anesthetized with dexmedetomidine (30 mcg/kg, IM) (Zoetis Animal Health, Parsippany, NJ, USA) combined with ketamine (25 mg/kg, IM) (Zoetis Animal Health, Parsippany, NJ, USA), intubated and maintained under isoflurane (Vet One, Boise, ID, USA) general inhalant anesthesia (1-2% + 1-2 L/min medical grade O2). Systemic analgesia consisted of a single pre-operative dose of buprenorphine HCl (0.03 mg/kg, SC) (Hospira Pharmaceuticals, Lake Forest, IL, USA) followed by two doses of sustained release Buprenorphine-SR LAB (0.12 mg/kg SC every 72 hr) (Wildlife Pharmaceuticals, New Windsor, CO, USA). Peri-operative fluid and drug administration included isotonic saline (3 mL/kg/hr, IV, intra-op.) (Medline Industries Inc, Northfield, IL, USA), cefazolin (22 mg/kg, IV, once, pre-op.) (Westward Pharmaceuticals, Eatontown, NJ, USA) and maropitant citrate (Cerenia®, 1mg/kg, SC, SID x 5 days) Zoetis Animal Health, Parsippany, NJ, USA), Heart rate, respiratory rate, body temperature, end tidal CO2, SPO2, and EKG were monitored continuously and documented every 10 – 15 minutes during surgery. Thermal support (Bair Huggers) was used when indicated during surgery and initial recovery. Animals were monitored continuously for 1- 2 hours following estuation and the regaining of consciousness and at least twice daily thereafter through study termination. Daily evaluations included local and systemic pain scoring, surgical site inspection, neurologic, musculoskeletal and gastrointestinal functional assessments.
In-vivo animal experiments
All In-vivo animal experiments were reviewed and approved by the Ibex Preclinical Research Institutional Animal Care and Use Committee (Ibex Preclinical Research, Inc., Logan, UT) in compliance with the Animal Welfare Act, supporting federal statutes, regulations and principles as stated in the Guide for the Care and Use of Laboratory Animals[52]. 33 skeletally mature specific pathogen free (SPF) 7-month-old nulliparous, female New Zealand White (NZW) rabbits (6.5-7.8 kg at study initiation) (Western Oregon Rabbit Company, Philomath, OR, USA) were randomly assigned to either cranial defect (n=15) or spinal fusion studies (n=18). Rabbits were singly housed per the study parameters in 68.6 W x 68.6 D x 45.0 H cm primary enclosures (Allentown, Inc. PA, USA) equipped to provide ad libitum chlorinated water and enriched with stainless steel bells and polypropylene dumbbells (Bioserv, Flemington, NJ, USE). In addition to ad libitum pelleted feed (Teklad Global High Fiber Rabbit Diet [2031], Envigo, USA), rabbits received daily fresh produce (e.g. lettuce, apples,), orchard grass hay (Ibex-sourced), and commercially available treats (Bioserv, Flemington, NJ, USA). Animal holding rooms were maintained on a 12:12 h light:dark cycle at 15–21 C° with a relative humidity of 45–65%.
All animals underwent a 10-14-day acclimation period at the research facility prior to study initiation. Animals were examined by the preclinical vivarium husbandry and technical staff at least twice daily seven days a week from acclimation through study termination. Clinical and experimental evaluations were reviewed daily by the Attending Veterinarian or designee.
Cranial defect model
Animals were placed in ventral recumbency for surgery to create two paramedial, 8 mm (outer diameter), bicortical parietal bone defects. A midline skin incision was made from the nasofrontal. The periosteum was incised and reflected away from the underlying parietal bones. Defects were created using an 8 mm short trephine drill and contra angle handpiece on a single implant motor system (Treph-Short-8, Motor-Single-20-1; Salvin Dental Specialties, Inc, Charlotte, NC, USA) with continuous isotonic saline (Abbott Laboratories, North Chicago, IL, USA) irrigation. Care was taken to avoid damage to the underlying dura or microvasculature.
Animals were randomized using a card-based technique to receive 1) split calvarial autografts (SCA) (n=6), 2) AHBC (n=6), or 3) DBM+BMP-2 (n=6). SCA was generated using a straight 2- mm osteotome (N4302, Bausch Health Companies Inc. Rochester, NY) to separate the inner and outer cortices of bone harvested from the defect. The outer cortex was re-implanted into the defect. AHBC was generated intra-operatively in accordance with manufacturer’s processing methods using the entire quantity of parietal bone harvested from the defect. The resultant AHBC test article was implanted back into the defect. The human DBM (DGC 1/8; OraGRAFT® DBM, Life Net Health, Virginia Beach, VA) and BMP- 2 (CYT-261; ProSpec, New Brunswick, NJ) was prepared through combination of 0.5 mL DBM and 5 micrograms BMP-2. The resultant mixture was implanted into the defect. One of the defects remained open (untreated) across experimental groups to serve as an internal negative control. Periosteum was re-opposed over both defects with 4-0 suture (Monocryl®, Ethicon, Inc., Somerville, NJ). Skin was reopposed with 3-0 suture (Biosyn™, Covidien, Dublin, Ireland).
Posterolateral lumbar spinal fusion model
Animals were placed in ventral recumbency for bilateral iliac crest harvest and transverse lumbar spinous process fusion. Right and left iliac crests and 0.5 cm of adjacent ilium body were resected with Beyer Rongeurs (160002-18; Fine Science Tools, Foster City, CA, USA) yielding 1.6 – 1.8 grams of cortico-cancellous bone per crest divided between the right and left L4-L5 PLF sites (AHBC and ABG treatment groups). A dorsal midline skin incision was created from lumbar vertebral body 3 to 6. Bilateral paraspinal incisions were made to expose and reflect the erector spinae muscles and fasciae covering the dorsal and lateral aspects of the right and left L4- L5 transverse processes. A 1.2 mm step drill urban decorticating bur and straight handpiece on a single implant motor system (Lindemann-su Motorsingle- 20-1) Salvin Dental Specialties, Inc., Charlotte, NC, USA) was used at 300 rpm to decorticate the dorsal surfaces of the transverse processes of L4 and L5 extending approximately 2 cm laterally from the transverse process (TP)/pars interarticularis junction. Animals were randomized using a card-based technique to receive autologous bone grafts (ABG) (n=6), AHBC (n=6), or DBM+BMP-2 (n=6). Human DBM (DGC 1/8; OraGRAFT® DBM, Life Net Health, Virginia Beach, VA, USA) and BMP-2 (CYT-261; ProSpec, New Brunswick, NJ) was prepared through combination of 2.5 mL DBM and 10 micrograms BMP-2. ABG was derived from iliac crest bone morselized by surgeons intra-operatively using Beyer Rongeurs (160002-18; Fine Science Tools, Foster City, CA, USA). AHBC was generated intraoperatively in accordance with manufacturer’s processing methods using the entire quantity of harvested iliac crest bone. Test article was equally divided between right and left L4-L5 processes to bridge from the cranial aspect of the L4 TP to the caudal aspect of the L5 TP. Paraspinal muscle, fascia and skin were closed in three layers with absorbable 3-0 suture (Biosyn™, Covidien, Dublin, Ireland).
Necropsy/Macroscopic Evaluation
Animals were euthanized on POW 8 in accordance with the American Veterinary Medical Association (AVMA) Guidelines on Euthanasia [53]. Briefly with animals under general inhalant anesthesia following the final CT session and intravenous auricular vein catheter was placed for administration of veterinary medical grade pentobarbital-based euthanasia solution (Vet One, Boise, ID, USA) dosed according to manufacturer instructions. Full necropsies and manual palpation (spinal study only) were conducted on all animals according to standard operating procedures under the supervision of the principal investigator (PI) and Attending Veterinarian. The entire skull or thoracolumbar segment of spine was carefully excised en-bloc. Soft tissues and fascia were carefully removed from around each experimental site after the skull and spine were explanted. Each grafted site was examined for test article migration, infection, and soft tissue abnormalities.
CT scans
Longitudinal post-operative CT (Vimago, Epica Medical Innovations, San Clemente, CA) was performed immediately postsurgery (POD 0) and again at 4, and 8 weeks (cranial defect model) and POD 0, 2, 4, 6, 8, 10, and 12 weeks (spinal fusion model). The following settings were used for both models: 60 mA, 80 kV, 7 ms, time: 32 seconds, resolution: 200 μm. A Quantum GX2 (PerkinElmer, Waltham, MA) instrument was used to image all ex-vivo rabbit crania and spine specimens. Each crania specimen was imaged at 70 kV, 88 μA, FOV 36 mm, voxel size 90 μm, Al 0.5 CU 1.0 filter for 14 minutes to achieve best resolution. Spines were imaged at 90 kV, 88 μA, FOV 86 mm, voxel size 172 μm, Al 0.5 CU 1.0 filter for 4 minutes on the same machine.
Images were exported as DICOM file format then imported and analyzed with Analyze software version 12.0 (AnalyzeDirect, Overland Park, KS). The trabecular and cortical bone mineral densities (BMD) were determined using one phantom (25 mm QRM BMD phantom, QRM Quality Assurance in Radiology and Medicine GmbH, Möhrendorf, Germany) with known densities of 50 mg/cm3, 200 mg/cm3, 800 mg/cm3, and 1200 mg/cm3 of hydroxyapatite. Thresholds were set at were set at 539 Hounsfield units (HU), for crania and 220 HU for spine, based off visual analysis. Statistical analysis was performed using GraphPad Prism 7.04 (GraphPad Software, San Diego, CA). A Dunnett’s multiple comparison test was used to determine statistically significant differences among groups. Either the native or untreated groups were used as the control in the Dunnett’s multiple comparison test.
For the spinal studies, statistical analysis was performed using R statistical programming language (R Foundation for Statistical Computing, Vienna, Austria). The region of interest was selected via two raters. A Dunnett’s multiple comparison test was used to determine statistical significance (p ≤ 0.05). All measurements were made comparing all samples to autograft. To determine frequency of fusion two raters twisted and flexed ex-vivo rabbit spine specimen. Values were recorded and processed in GraphPad Prism version 7.04 (GraphPad Software, San Diego, CA).
Raman spectroscopy
A confocal Raman microscope (Raman DXR Microscope, Thermo Fisher Scientific, Waltham, MA) with a 10x objective and a laser wavelength of 785 nm (28 mW laser power) was used to collect spectra. A 25-um slit aperture was used to collect a spectral range between wavenumbers 500-3500 cm-1. The estimated resolution was 2.3-4.3 cm-1. Spectral data was collected using an exposure of 1 s with a signal to noise ratio of 300 to ensure the collected spectra represent the bulk material. For surface point scans, a total of 2-5 spectra were collected from arbitrary positions across the top surface of the defect. For surface line scans, 6 spectra were collected with 200 μm spacing between each point of collection. In addition to point and line scans, cross sectional area scans were collected for each subject defect. Area scans consisted of full thickness cross sections covering an area between 3-15 mm2 with 100-320 points of collection.
Raman spectroscopy analysis was performed using software for Dispersive Raman (OMNIC v.32, Thermo Fisher Scientific, Waltham, MA) to remove background fluorescence from all surface point scan spectra using 6th order polynomial baseline fitting. Surface point spectra collected from each specimen were normalized and averaged to represent an individual subject. Overall group averages were calculated using average spectra from each individual subject within the group. Chemigrams for cross sectional area scans were created using ranges 950-965 cm-1 (hydroxyapatite) and 880-840 cm-1 (collagen).
Mineral-to-collagen Raman peak ratios were calculated and compared to ratios of native rabbit crania and control defect [54]. Surface point scans were utilized for calculation of peak ratios.
Macros Basic for the software (OMNIC v.32, Thermo Fisher Scientific, Waltham, MA) was used to determine the uncorrected peak heights of amide I (1663 cm-1), amide III (1253 cm-1), B-type carbonate (1070 cm-1), proline (856 cm-1), and ν1 phosphate (961 cm-1). Find nearest peak was enabled on the macro to ensure the correct peak was recorded. Peak ratios consisted of phosphate to amide I, phosphate to amide III, phosphate to proline, and carbonate to phosphate.
Mechanical Testing
Samples were tested for indentation strength using an electronic universal testing machine (UTM) with 1 kN load capacity (Universal Testing Machine, Instron, Norwood, MA) at a constant crosshead velocity of 1 mm/min until break point was reached. One indentation location was used on each specimen and 3 indentations were carried out at each location. Load was applied along the transverse axis of the bone. Load and displacement values were recorded at 0.1 s intervals during testing.
The torsion tests were conducted with an electronic UTM with 25 nm torsional load capacity (Instron, Norwood, MA) to a total displacement of 5 degrees at a rate of 10 deg/s. Torque versus rotational displacement plots were obtained for each test group (n=6), and torsional stiffness was determined in the range of 20% to 90% of total displacement.
Scanning Electron Microscopy
Using an environmental scanning electron microscope (Zeiss Evo LS 10, Carl Zeiss AG, Oberkochen, Germany), images were taken of subjects. Using a back-scatter detector (High Definition Back-Scatter Detector (HDBSD), Carl Zeiss AG, Oberkochen, Germany), with medium gain and all 5 sections of the detector on, the samples were imaged at 30X, 60Pa, 20Kv, using Atlas 5 image collection software. The HDBSD gives qualitative image contrast through detection of the electrons in samples where a brighter region is more electron dense, higher atomic weight, and a darker region is less electron dense.
Multiphoton Microscopy
Second-harmonic imaging was performed using a multiphoton confocal microscope equipped with a two-photon laser and a 10 x 0.40 NA objective (Leica SP8, Leica Microsystems, Wetzlar, Germany). Excitation was done with an 880 nm tuned Coherent Chameleon Vision II laser. The second-harmonic emission signals were detected using a HyD detector (Leica HyD, Leica Microsystems, Wetzlar, Germany) at 430 nm to 450 nm and converted to TIF format using software (Leica application Suite X, Leica Microsystems, Wetzlar, Germany). SEM and MPM were performed to examine the bone ultrastructure across different treatments in cranial defects and spinal fusion masses.
Compound Macro Imaging
Compound macro-imaging was performed using a stereo microscope (Leica M205 FA, Leica Microsystems, Wetzlar, Germany). Samples were viewed with a 0.63x planapo lens at 1, 2, 5, and 10x zoom and images were collected using a camera (Leica DFC7000 T, Leica Microsystems, Wetzlar, Germany).
Confocal Microscopy
Confocal fluorescent imaging was performed using a single photon confocal microscope (Leica TCS SP8, Leica Microsystems, Wetzlar, Germany). Samples were imaged with 10x 0.40 NA objective. Samples labeled with probes (NucBlue: R37605, Thermo Fisher Scientific, Waltham, MA, Osetoimage: PA-1503, Lonza, Basel, Switzerland, and ActinRed 555: R37122, Thermo Fisher Scientific, Waltham, MA) were visualized using 405 (Diode), 488 (Argon), 514 (Diode), and 633 (HeNe) laser lines and signals were detected using a combination detection system (Leica HyD and PMT, Leica Microsystems, Wetzlar, Germany). Images were viewed and converted to TIF format using software (Leica application Suite X, Leica Microsystems, Wetzlar, Germany).
Histology
Excised tissues were fixed in 10% neutral buffered formalin, decalcified in Mol-Decalcifier (Milestone Medical, Kalamazoo, MI, USA) and infiltrated with paraffin using a Sakura Tissue- Tek VIP 6 AI tissue processor. Paraffin-embedded samples were sectioned to 3.5-micron thickness and mounted on positively charged slides prior to Hematoxylin and Eosin (H&E) staining on a Sakura Tissue- Tek Prisma Plus Automated slide stainer and Masson’s Trichrome (MT) staining using a StatLab staining kit (StatLab Medical Products, McKinney, TX, USA) according to the manufacturer’s instructions.
RNA extraction
Rabbit. Native bone and AHBC generating from cranium or spine were collected from five rabbits. Tissue was immediately stored in stabilization media (Catalog #: 76405, Allprotect Tissue Reagent, Qiagen Inc., Hilden, Germany), then held at 4°C for 24 hours before being held at -80°C for storage until RNA extraction was performed. Lysis of tissue was performed with a tissue homogenizer (PowerLyzer, Qiagen Inc., Hilden, Germany) for two cycles of 45 seconds at 3500 rpm with a 30 second dwell time between cycles. RNA was purified from the resulting tissue lysate using a purification kit (Catalog #: 74134, RNEasy Plus Universal Mini Kit, Qiagen Inc., Hilden, Germany).
Human. Bone from one individual was obtained from a tissue service (Table 5: Hu0081, Donor Connect, Salt Lake City, UT). A portion was processed to AHBC. Triplicate 100 mg native bone and AHBC samples were flash frozen in liquid nitrogen and stored at -80°C until RNA extraction. Lysis of tissue was performed with a tissue homogenizer (PowerLyzer, Qiagen Inc., Hilden, Germany) using one 10 second cycle at 3500 rpm. RNA was purified from the resulting tissue lysate using a purification kit (Catalog #: 74134, RNEasy Plus Universal Mini Kit, Qiagen Inc., Hilden, Germany).
RT2 profiler array
RNA was quantified (Catalog #: ND-LITE, Nanodrop Lite (rabbit) or Catalog #: Q33226, Qubit (human), Thermo Fisher Scientific, Waltham, MA) from rabbit and human tissue samples. One native rabbit sample did not produce good RNA yield and was omitted from further analyses.
Eight hundred (rabbit) or 400 (human) nanograms of RNA were reverse transcribed to cDNA (RT2 First Strand Kit, Qiagen Inc., Hilden, Germany). Resulting cDNA was used as the template for RT2 PCR Profiler plates which were run according to manufacturer instructions (Qiagen Inc., Hilden, Germany) on a real-time PCR system (QuantStudio 12K Flex or QuantStudio 3, Thermo Fisher Scientific, Waltham, MA). Osteogenesis (rabbit), angiogenesis (human and rabbit), and wound healing (human and rabbit) arrays were run in triplicate (angiogenesis) and quadruplicate (wound healing).
Proteomic analysis of rabbit and human AHBC
Proteomic analysis was carried out on rabbit cortico-cancellous parietal bone pre- and post- processing, based on MS2-based TMTlabeling as previously described [55]. Briefly, pre- and post- processing bone specimens (n = 4) were homogenized in a PowerLyzer (Qiagen Inc., Hilden, Germany) as described above. Homogenates were centrifuged at 16,100G for 5 minutes at room temperature, with the supernatant assayed for protein content by Pierce 660 nm Assay (Catalog #: 22662, Thermo Fisher Scientific, Waltham, MA). 100 μg proteins were reduced, alkylated, and serially digested with LysC and trypsin. Desalted peptides were labeled with the 126, 127N, 127C, 128C, 129N, 129C, 130C, and 131 reagents from TMT 10plex (Catalog #: 90111, Thermo Fisher Scientific, Waltham, MA), combined, fractionated at pH ~9 by ion-exchange chromatography, and analyzed by reversed-phase C18-based chromatography by LC-MS/MS on a Q Exactive HF-X (Thermo Fisher Scientific, Waltham, MA). TMT data analysis was performed in Proteome Discoverer 2.3 software (Thermo Fisher Scientific, Waltham, MA). Protein identification was done by the SequestHT and Percolator algorithms, with searching carried out against rabbit protein sequences with the taxonomy ID 9986 (Uniport Knowledge Base, April 2019). In total, 2735 proteins were identified (false-discovery rate <1%) and quantified. Protein abundance values were normalized to pre-defined housekeeping proteins (β - tubulin, actin, and histone H4) in the individual samples. 741 proteins exhibited ≥ 1.5-fold difference between native bone and AHBCtreated bone in at least one of the 4 animals; the Log10-transformed abundance values were subjected median-normalization followed by two- dimensional hierarchical clustering. The 741 proteins also served as input for principal component analysis by row-wise estimation. To identify biological processes over-represented by proteins with abundance changes, a total of 20 Gene Ontology Biological Processes terms (as annotated in Uniport) were found to be represented by ≥ 4 proteins. The abundance values of the constituent proteins were summed, followed by two-dimensional hierarchical clustering as described above.
Proteomic analysis was carried out on cadaveric human donor tissues (n = 3) pre- and post- processing as was on rabbit tissues, with the exception that the six samples were labelled with the TMT 6plex assay kit (Catalog #: 90066, Thermo Fisher Scientific, Waltham, MA). Log10- transformed abundance values were subjected to twodimensional hierarchical clustering. Gene ontology analysis was performed using the DAVID Bioinformatics Resources v6.8 [56].
Statistical Analyses
Bone mineral density data and cranial indentation modulus data was analysed using statistical software (GraphPad Prism v 7.04, GraphPad Software, San Diego, CA). Raw values were analysed first using one-way ANOVA (p ≤ 0.05), then using Tukey’s multiple comparison test. Data is expressed as mean ± standard deviation. Histograms were analysed using analysis software (ImageJ, National Institutes of Health, Bethesda, MD) and are reported as mean ± standard deviation.
qPCR data was analysed using statistical software (R version 3.5.2, R Foundation for Statistical Computing, Vienna, Austria). Undetermined values on each array were given the mean value for the respective group (AHBC or native) if two or more samples had detectable values. If less than two samples had detectable values the undetected values were given a value of 40 (max number of cycles run). The data was quantile normalized. Hierarchical clustering and principal component analyses were performed using the HTqPCR package, under suggested conditions [57].
Sample Size
A power analysis based on a fixed effects, repeated measures ANOVA algorithm determined that a sample size of n= 6 per experimental group is indicated to observe a significant impact of treatment on cranial or spinal defect site morbidity as assessed by ex vivo mechanical testing and computed tomography with an effect size of 0.4 at a power level of 0.95 and alpha of 0.05. Data variability and effect size is comparable or reduced for molecular data acquired for assessment of secondary outcomes, and thus a total group sample size of n=6 rabbits per group is sufficient for all planned analyses using standard tissue harvesting and partitioning practices.
Animal age and body mass were tightly controlled to avoid any confounding influence of age or weight on the results of the study. All ex-vivo measurements including indentation modulus and imaging analysis were conducted by the contributing author blinded to treatment group. All molecular, proteomic data acquisition and analysis on tissue samples acquired were likewise analyzed in a blinded fashion. Indications for stoppage of data collection on a given animal prior to the cohort endpoint was tracked by the principal investigator in consultation with the Attending Veterinarian or designee as necessary for animal wellness according to IACUC approved criteria.
Results
AHBC Viability Ex-Vivo
AHBC retains the relevant endogenous osteogenic and supportive cellular populations found in native bone. Alamarblue assay was performed to evaluate the viability of these populations within AHBC created from fresh goat iliac bone during short-term storage at 4C over 10 days. Ratio of Day 1 fluorescence is shown in (Figure 14) and demonstrates the change over time for AHBC. AHBC was significantly different than dead controls on all days assessed (P< 0.05) and not significantly different than native tissues (Figure 14). Three technical replicates were used from each biological sample at each timepoint unless tissue volume was limited, in which case 2 technical replicates were used.
AHBC and ABG treated fusion masses are structurally and mechanically similar
Raman spectra of AHBC and ABG treated fusion masses had the mineral peak at 957 cm-1 while the DBM treated group did not have that peak in the spectra of the fusion mass. ABG and AHBC display extensive mineral deposition as well as complete fusion, while DBM+BMP2 appears to have little osteogenesis and partial fusion (Figure 15).
Figure 15: Representative intervertebral samples from each group imaged with high resolution digital single lens reflex (DSLR) photography (top row) and scanning electron microscopy (SEM) (bottom row). All samples were imaged ex vivo post-operative week 8 and demonstrate that AHBC-generated osseous spinal ultrastructure is analogous to autograft. Spinous process on right for AHBC and autograft, not present for DBM+BMP2.
Ex-vivo bone indentation was measured on detectable spinal fusion masses for AHBC generated tissue, DBM+BMP2 generated tissue, and ABG generated tissue to assess the stiffness of fusion masses (Figure 12). There was no significant difference in indentation modulus values between any groups, however none of the DBM+BMP2 animals had complete fusion masses or masses were considerably smaller and were not uniform or completely connected between the vertebral bodies. Additionally, ABG and AHBC groups have more similar values than DBM+BMP2; ABG vs. DBM+BMP2: 37.001 MPa ± 27.189 vs. 8.137 MPa ± 6.551, p = 0.123; ABG vs. AHBC: 37.001 MPa ± 27.189 vs. 38.452 MPa ± 49.627, p = 0.952; AHBC vs. DBM+BMP2: 38.452 MPa ± 49.627 vs. 8.137 MPa ± 6.551, p = 0.347. Raman spectra comparison of AHBC generated tissue, ABG generated tissue, and DBM+BMP2 tissue was performed to observe collagen and hydroxyapatite levels (Figure 12). ABG and AHBC spectral fingerprints of collagen and hydroxyapatite are comparable, while DBM+BMP2 has lower levels of both
Acknowledgements
All the authors were paid employees of Polarity TE MD, Inc. during the completion of the study. The authors thank Devin T. Miller and colleagues at IBEX Research for their assistance in preclinical studies and Amanda Stump for assistance in the literature review.
Ethical approval
All In-vivo animal experiments were reviewed and approved by the Ibex Preclinical Research Institutional Animal Care and Use Committee (Ibex Preclinical Research, Inc., Logan, UT) in compliance with the Animal Welfare Act, Regulations, and Care Policies
Conflict of Interest Statement
All the authors are paid and employed by Polarity TE MD, Inc.
References
- Fernandez de Grado G, Keller L, Idoux-Gillet Y (2018) Bone substitutes: a review of their characteristics, clinical use, and perspectives for large bone defects management. J. Tissue Eng. 9.
- Desilva GL, Lou M, Greenfield KH (1997) Autogenous Iliac Crest Bone Graft:Complications and Functional Assessment. Clin Orthop Relat Res.399:76– 81.
- Herford AS, Dean JS (2011) Complications in Bone Grafting. Oral Maxillofac. Surg. Clin. North Am.23:433–442.
- Kinaci A, Neuhaus V, Ring D (2014) Trends in Bone Graft Use in the United States. Orthopedics 37:e783-e788.
- Dumas JE, Zienkiewicz K, Tanner SA (2010) Synthesis and Characterization of an Injectable Allograft Bone/Polymer Composite Bone Void Filler with Tunable Mechanical Properties.Tissue Eng. Part A 16: 2505-2518
- Mankin HJ, Hornicek FJ, Raskin KA (2005) Infection in massive bone allografts. Clin. Orthop. Relat.Res.432:210–216.
- Martin GJ, Boden SD, Titus L, Scarborough NL (1999) New Formulations of Demineralized Bone Matrix as a More Effective Graft Alternative in Experimental Posterolateral Lumbar Spine Arthrodesis.Spine:637–645.
- Gruskin E, Doll BA, Futrell FW (2012) Demineralized bone matrix in bone repair: History and use. Adv.Drug Deliv.Rev.64:1063–1077
- Gruskin E, Doll BA, Futrell FW (2012) Demineralized bone matrix in bone repair: History and use. Adv. Drug Deliv.Rev.64:1063–1077.
- Peterson B, Whang PG, Iglesias R (2004) Osteoinductivity of Commercially Available Demineralized Bone Matrix Preparations in a Spine Fusion Model. J.Bone Jt.Surg.86:2243–2250.
- Mckee MD (2006) Management of Segmental Bony Defects :The Role of Osteoconductive. J.Am.Acad.Orthop.Surg. 14:163–167.
- Massague J (1990) The transforming growth factor-beta family. Annu. Rev. Cell Biol. 6:597–641.
- Chen B, Lin H, Wang J (2007) Homogeneous osteogenesis and bone regeneration by demineralized bone matrix loading with collagen-targeting bone morphogenetic protein-2. Biomaterials 28:1027–1035.
- Zhou N, Li Q, Lin X (2016) BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell Tissue Res. 336:101– 111.
- Vaidya R, Carp J, Sethi A (2007) Complications of anterior cervical discectomy and fusion using recombinant human bone morphogenetic protein-2. Eur. Spine J.16:1257– 1265.
- Carragee EJ, Hurwitz EL, Weiner BK (2011) A critical review of recombinant human bone morphogenetic protein-2 trials in spinal surgery: Emerging safety concerns and lessons learned. Spine J.11:471–491.
- James AW, LaChaud G, Shen J (2016) A Review of the Clinical Side Effects of Bone Morphogenetic Protein-2. Tissue Eng. - Part B Rev.22: 284–29
- Epstein N. 2013. Complications due to the use of BMP/INFUSE in spine surgery: The evidence continues to mount. Surg. Neurol. Int. 4: S343–S352
- Woo EJ (2012) Adverse Events Reported After the Use of Recombinat Human Bone Morphogenetic Protein 2. J. Oral Maxillofac. Surg.70:765–767.
- Wancket LM (2015) Animal Models for Evaluation of Bone Implants and Devices: Comparative Bone Structure and Common Model Uses. Vet. Pathol. 52:842–850.
- Stübinger S, Dard M (2013) The Rabbit as Experimental Model for Research in Implant Dentistry and Related Tissue Regeneration. J. Investig. Surg.26:266-82.
- Leventis M, Fairbairn P, Mangham C (2018) Bone healing in rabbit calvaria defects using a synthetic bone substitute: A histological and micro-CT comparative study. Materials (Basel).11
- (2017) ASTM International Standard Guide for in vivo Evaluation of Rabbit Lumbar Intertransverse Process Spinal Fusion Model.F3207-17
- (2012) ASTM International Standard Guide for Pre-clinical in vivo Evaluation of Spinal Fusion.F2884-12
- (2016) ISO Biological evaluation of medical devices - Part 6: Tests for local effects after implantation.10993–6
- Leary S, Underwood W, Anthony R, Cartner S (2013) AVMA Guidelines for the Euthanasia of Animals:2013 Edition
- Cottrell JM, Van Der Meulen MCH, Lane JM, Myers ER (2006) Assessing the stiffness of spinal fusion in animal models. HSS J.2:12
- Bi X, Patil CA, Lynch CC (2011) Raman and mechanical properties correlate at whole bone- and tissue-levels in a genetic mouse model. J. Biomech.44:297-303
- Barnes AM, Carter EM, Cabral WA (2010) Lack of Cyclophilin B in Osteogenesis Imperfecta with Normal Collagen Folding.N Engl J Med.362:521–528.
- Elshahat A, Shermak MA, Inoue N (2004) The Use of Novabone and Norian in Cranioplasty: A Comparative Study. J. Craniofac.Surg. 15:483-489
- Wang JC, Alanay A, Davies M (2007) A comparison of commercially available demineralized bone matrix for spinal fusion. Eur. Spine J.16:1233–1240.
- Movassaghi K, Ver Halen J, Ganchi P, Amin-Hanjani S (2006) Cranioplasty with Subcutaneously Preserved Autologous Bone Grafts. Plast. Reconstr. Surg. 117:202–206.
- Buser Z, Brodke DS, Youssef JA (2018) Allograft Versus Demineralized Bone Matrix in Instrumented and Noninstrumented Lumbar Fusion: A Systematic Review. Glob. Spine J.8:396–412
- Provaggi E, Capelli C, Leong JJH, Kalaskar DM (2018) A UK-based pilot study of current surgical practice and implant preferences in lumbar fusion surgery. Med. 97:11169.
- Gopalakrishnan MS, Shanbhag NC, Shukla DP (2018) Complications of decompressive craniectomy. Front. Neurol.9:5–7.
- Lee SS, Jang JH, Kim KS (2009) Failure of bone regeneration after demineralized bone matrix allograft in human maxillary sinus floor elevation. Basic Appl. Pathol. 2:125–130.
- Martin KD, Franz B, Kirsch M (2014) Autologous bone flap cranioplasty following decompressive craniectomy is combined with a high complication rate in pediatric traumatic brain injury patients. Acta Neurochir.156:813–824.
- Boden SD (2002) Overview of the Biology of Lumbar Spine Fusion and Principles for Selecting a Bone Graft Substitute.27:26–31.
- McEntire BJ, Bal BS, Rahaman MN (2015) Ceramics and ceramic coatings in orthopaedics. J. Eur. Ceram. Soc.35:4327-4369
- Rossi F, Santoro M, Perale G (2015) Polymeric scaffolds as stem cell carriers in bone repair. J. Tissue Eng. Regen. Med. 9:1093-1119
- Hasan A, Byambaa B, Morshed M (2018) Advances in osteobiologic materials for bone substitutes. J. Tissue Eng. Regen. Med.12:1448-1468.
- Yang W, Cao Y, Zhang Z (2018) Targeted delivery of FGF2 to subchondral bone enhanced the repair of articular cartilage defect. Acta Biomater. 69:170–182
- Arora M, Chan EKS, Gupta S, Diwan AD (2013) Polymethylmethacrylate bone cements and additives: A review of the literature. World J. Orthop.4:67-74.
- Ronga M, Fagetti A, Canton G (2013) Clinical applications of growth factors in bone injuries: Experience with BMPs. Injury. 44
- Faundez A, Tournier C, Garcia M (2016) Bone morphogenetic protein use in spine surgery—complications and outcomes: a systematic review. Int. Orthop.40:1309-19
- Nguyen V, Meyers CA, Yan N (2017) BMP-2-induced bone formation and neural inflammation. J. Orthop.14:252-256.
- Tannoury CA, An HS (2014) Complications with the use of bone morphogenetic protein 2 (BMP-2) in spine surgery. Spine J.14:552-559
- Bach DH, Park HJ, Lee SK (2018) The Dual Role of Bone Morphogenetic Proteins in Cancer. Mol. Ther- Oncolytics.8:1-13
- Chen SS, Fitzgerald W, Zimmerberg J (2007) Cell-Cell and Cell-Extracellular Matrix Interactions Regulate Embryonic Stem Cell Differentiation. Stem Cells.25:553-61.
- Alford AI, Kozloff KM, Hankenson KD (2015) Extracellular matrix networks in bone remodeling. Int. J. Biochem. Cell Biol. 65:20-31
- Smith LR, Cho S, Discher DE (2018) Stem cell differentiation is regulated by extracellular matrix mechanics. Physiology.33:16-25
- National Research Council.(2011). Guide for the Care and Use of Laboratory Animals: Eighth Edition. Washington, DC: The National Academies.
- Leary S, Underwood W, Anthony R, Cartner S (2013) AVMA Guidelines for the Euthanasia of Animals: 2013 Edition.
- Bi X, Patil CA, Lynch CC, Pharr GM (2011) Raman and mechanical properties correlate at whole bone- and tissue-levels in a genetic mouse model. J Biomech. 44:297-30
- Hogrebe A, von Stechow L, Bekker-Jensen DB (2018) Benchmarking common quantification strategies for large-scale phosphoproteomics. Nat Commun.9:1045.
- Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res.37:1-13
- Dvinge H (2019) HTqPCR high-throughput qPCR analysis in R and Bioconductor.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi