Research Article, J Vet Sci Med Diagn Vol: 6 Issue: 5
Investigation of Infectious Bronchitis Virus Strains by Real Time RT-PCR and S1 Gene Sequencing in Broilers
Engin Alp Onen1* and Yakut Ozgur2
1Kocak Farma ve Ilac San. Tic. AS Biotechnology Department, Turkey
2Istanbul University, Faculty of Veterinary Medicine, Department of Microbiology, Turkey
*Corresponding Author : Engin Alp Onen, PhD
Kocak Farma ve Ilac San. Tic. AS Biotechnology Department, Istanbul University, Turkey
E-mail: alponen@gmail.com
Received: October 20, 2017 Accepted: November 13, 2017 Published: November 17, 2017
Citation: Onen EA, Ozgur Y (2017) Investigation of Infectious Bronchitis Virus Strains by Real Time RT-PCR and S1 Gene Sequencing in Broilers. J Vet Sci Med Diagn 6:5. doi: 10.4172/2325-9590.1000243
Abstract
In this study, detection of infectious bronchitis virus (IBV) from chicken tracheal swab samples by real time RT-PCR, which was reported as a rapid and sensitive virus detection method, was aimed. Phylogenetic relations in different field and variant strains of IBV by sequencing of hyper variable regions (HVRs) of S1 gene of IBV was determined. Tracheal swabs were taken from 120 broiler chickens with respiratory signs, randomly. Samples were inoculated into the allantoic fluids (AF) of SPF embryonated chicken eggs. RNAs were extracted by AF. RNAs were translated to ss cDNAs. Real time RT-PCR was performed with double labelled Taqman probe, IBV 5 GU391 and IBV5 GL533 primer pairs. Positive results were obtained in 33 samples of 120 (27.5 %) by real time RT-PCR. cDNAs from positive samples were used for amplification HVRs (Hyper Variable Regions) in S1 gene region with C2U and Ag0723 primers. Samples were amplified by S1 5’mod and CK2 primers, too. RT-PCR products were sequenced directly. Sequencing results have been compared with NCBI’s nucleotide database. Sequencing results were matched to some variant IBV strains in gen banks database at the rate of 72-86 %. These results support that we are faced with variant IBV strains. Also, we observed a lot of SNP mutations in alignment. This study shows that birds can be infected by IBV variant strains at rate of 27.5 %, although they were vaccinated by classic type vaccine. These results demonstrate that vaccination by classical vaccine strains of IBV cannot provide enough cross protection against variant strains. Therefore, outbreaks of the disease still occur in vaccinated flocks. Also, we found these primers are not enough for all IBV field strains to be sequenced and classified. Thus, different primers should be developed and used primer pairs more than two. In this experiment, we found virus load is more in AF than swab samples because of IBV replication in SPF egg AF. We observed dwarfism and hemorrhages at 14 days old chick embryos
Keywords: IBV; Chicken; Real time RT-PCR; Sequencing; S1 gene; Hyper variable regions
Introduction
Infectious Bronchitis (IB) is an acute and considerably contagious upper respiratory disease which leads to an economic decrement in poultry industry affecting the performance of broiler and layer type of chickens.
Infectious bronchitis causes a decrease in weighting and feed conversion. Mix infections cause severe air sacks inflammation and increase rates of defections. Also E. Coli contributes to this mixed infection as a secondary pathogen. On the other hand, it leads to decrease the rate of quality and egg producing to layers [1-4].
IBV with large positive-sense RNA genome of 27.6 kb, is a prototype virus of Coronaviridae family and a member of genus Gammacoronavirus, subfamily Coronavirinae. It also has three main partial proteins such as spike glycoproteins (S), membrane proteins (M) and nucleocapsid proteins (N). Spike glycoproteins, S1 and S2 (520 and 625 composed of amino acids) consist of 2 components [2,5,6].
IBV has a great mutational changing capability. These mutations that which especially occurred in HVR (Hyper Variable Regions) of S1 gene cause to undergo a change in serotypes and strains. S glycoprotein is cleaved by proteolysis to S1 and S2 subunits. These subunits are responsible to binding of the IBV to the host cell. In Europe, it was found that the most common strains of IBV is relating to 793/B serotype. Also, this serotype was detected in Saudi Arabia, Japan, Sweden, Denmark, Italy, France and Argentina in 1997-1998. 793/B strain showed distinct clustering from earlier years although translated amino acid alternations were minimal [3,7-9].
Control of IBV is through use of live attenuated vaccines and inactivated vaccines, but the disease is difficult to control because different serotypes (including variant strains) of the virus do not cross-protect. Massachusetts type of live vaccines is generally used to prevent the disease [10].
There are many methods for diagnosis of acute IBV infection. Recently most relative diagnosis methods are the isolation of virus (VI), immune fluorescents assay (IFA), immune peroxidase assay (IPA) and reverse transcriptase polymerase chain reaction (RT-PCR). In serological definition of IBV infection, presence of IBV specific immunoglobulin M is detected. Hemagglutination inhibition (HI), agar gel precipitation (AGPT) and ELISA is most often used test in serological determination. Virus neutralization (VN) test is rarely used, because it is laborious and time consuming [11].
General molecular diagnosis of IBV is related to determination of N (Nucleocapsid) protein and protected region of replicas protein. Phylogenetic classification of IBV also depends on the analyzing of S1 (Spike 1) protein [12].
Materials and Methods
Samples
Samples were received from 13 different broiler poultry houses at times when respiratory signs were evident. Swabs were collected from the birds trachea and placed in the viral transport medium (VTM). VTM were stored at ice during the transport to the laboratory. Tubes that contain swab and VTM were vortex and swabs were discarded from VTM. Until the VTM was inoculated into SPF embryonating eggs, it was stored at – 70°C [10,13].
Virus multiplication
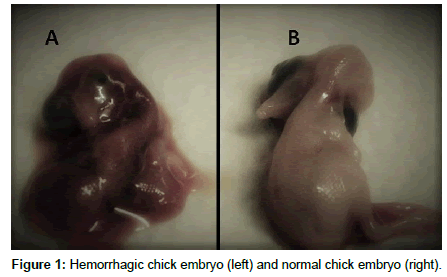
0.3 ml VTM was inoculated into allantoic fluid of 9- 11-day old SPF embryonated chicken eggs using by sterile 20-gauge syringe. Hole was closed by paraffin. Embryonated eggs were incubated at 38°C, 50 – 60 % relative humidity for 24 - 72 h. Inoculated eggs was candled at least once a day to identify eggs with dead embryos. Embryos that die within 24 hr were discarded as non spesific deaths caused by injury or bacterial contamination. All embryos which died beyond 24 h post infection were regarded as suspicious. If the inoculated eggs had lived embryos between 24 and 72 h, eggs were refrigerated for a minimum of 4 h to kill the embryo, harvested allantoic fluids were centrifuged at 3000 rpm for 10 min. and then allantoic fluids were harvested into screw capped tubes. Also we observed hemorrhage and dwarfism in embryos. Until the third passages or extraction of viral RNA, allantoic fluids were stored at – 70°C [9,13,14].
RNA extraction
Extraction of viral RNA from allantoic fluid of inoculated embryos was achieved with High Pure RNA Isolation Kit (ROCHE) following the manufacturer’s suggestions. Also, we carried out to extraction RNA from directly VTM.
Reverse transcription procedure
Reverse transcription of viral RNA was achieved with transcriptor High Fidelity cDNA synthesis kit (ROCHE) following the manufacturer’s suggestions. cDNAs were maintained at -20 0C until the following PCR and real-time PCR steps.
Real time PCR procedure
Primer probe mix was prepared with 5 μl of IBV5_GU391 (5’_-GCT TTT GAG CCT AGC GTT-3’_) forward primer, 5 μl of IBV5_GL533 (5’_-GCC ATG TTG TCA CTG TCT ATT G-3’_) reverse primer, 2 μl of IBV5_G (5’_-FAM CAC CAC CAG AAC CTG TCA CCT C-BHQ1-3’_) Taqman® dual-labeled probe [10] and 88 μl PCR grade water. 2 μl of primer probe mix was used for each of samples. For amplification of cDNAs, Light Cycler Taq Man Master kit (ROCHE) was used.
Master mix was prepared with adding 10 μl of enzyme into the 128 μl of reaction mix. For each of samples, 9 μl of PCR grade water, 2 μl of primer probe mix and 4 μl of master mix were mixing and after than PCR mix was obtained. For each of sample, 15 μl of PCR mix and 5 μl of samples were transferred into the Light cycler capillaries.
The reaction was conducted at Light Cycler 2.0 (ROCHE) at 95°C for 10 min; 45 cycles of 95°C for 10 s followed by 55 °C for 30 s and 72°C for 1s. In last step, real time procedure was ended at 40°C for 30 s. For each reaction, the cycle threshold (CT) number was determined corresponding to the PCR cycle number [10].
PCR for IBV S1 gene
Two different amplification processes were performed for sequencing of S1 gene. Two different primer sets were used for two different amplification of S1 region.
In first amplification of S1 gene HVRs (hyper variable regions), primer mix was prepared with 5 μl of C2U sense (5’-TGGTTGGCATTTACACGG (A/G) G-3’), 5 μl Ag0723 antisense (5’ -CACCYGCTGCTTCAACATC-3’) (16) primer and 90 μl PCR grade water. For each of reactions, PCR mix was prepared with 31.5 μl PCR grade water, 5 μl 10 x buffer + KCl, 4 μl 25 mM Mg2Cl, 0.4 μl 25 mM dNTP mix, 2 μl primer mix and 0.5 μl Taq polymerase. 10 μl templates (samples) were added to 43.4 μl PCR mix. PCR was conducted by heating to 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 30 secs, annealing at 50°C for 30 secs, and polymerization at 72°C for 7 min with a final elongation step of 10 min at 70°C using a thermal cycler (Eppendorf, MasterCycler). Amplification products were analyzed in a 1.0 % agarose gel.
Other amplification of S1 gene, we prepared primer mix with 5 μl of S1 5’mod forward (5’-TGAAAACTGAACAAAAGA-3’), 5 μl CK2 reverse (5’ -CNGTRTTRTAYTGRCA-3’) primer and 90 μl PCR grade water (19). For each of reactions, we prepared PCR mix with 28 μl PCR grade water, 5 μl 10 x buffer + KCl, 4 μl 25 mM Mg2Cl, 0.5 μl 25 mM dNTP mix, 2 μl primer mix and 0.5 μl Taq polymerase. 10 μl templates were added to 40 μl PCR mix. PCR was conducted by heating to 95°C for 2 min, followed by 45 cycles of denaturation at 95°C for 30 sec, annealing at 52°C for 30 sec, and polymerization at 68°C for 30 sec with a final elongation step of 12 min at 68°C using a thermal cycler. PCR products were maintained at +4°C [14]. Amplification products were analyzed in a 1.8% agarose gel.
Extraction and purification of PCR products from agarose gel for S1 sequencing
Extraction and purification of PCR products from agarose gel was performed with High Pure PCR Product Purification Kit (ROCHE) following the manufacturer’s suggestions.
S1 sequencing procedure
Extracted and purified RT-PCR products were sequenced directly with C2U, Ag0723, S1 5’mod forward and CK2 reverse primers by a sequencing device. Sequencing results have been blasted with 1000 bootstrap in NCBI’s nucleotide database.
Results
IBV specific embryonic lesions were observed such as dwarfing, stunting, curling and hemorrhages up to 3 passages of propagated embryos related to positive controls and some field samples. There were not any lesions in negative control groups.
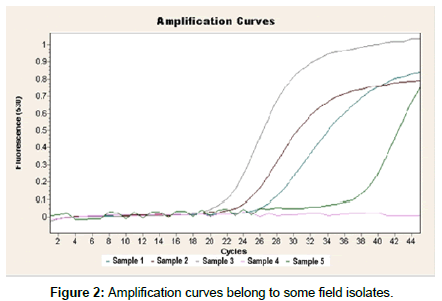
In this study, positive fluorescent signals were observed on 33 (27.5 %) samples of 120 from 13 different poultry houses. Positive results were obtained in positive control strains. No fluorescent signals were obtained in negative control groups. Also, positive signals were observed when real time RT-PCR was performed for direct sampling from VTM. Some real time PCR amplification results are shown in Figure 1.
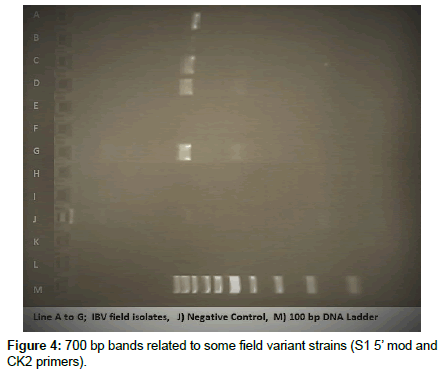
Approximately 450 bp bands were shown in electrophoresis corresponding to only M41 and H120 strains, but no bands were shown corresponding to field samples (Figure 2). In amplification with S1 5’ mod and CK2 primers, about 700 bp bands were shown in corresponding to 11 of field samples (Figure 3). No bands were shown in remained field samples (Figure 4).
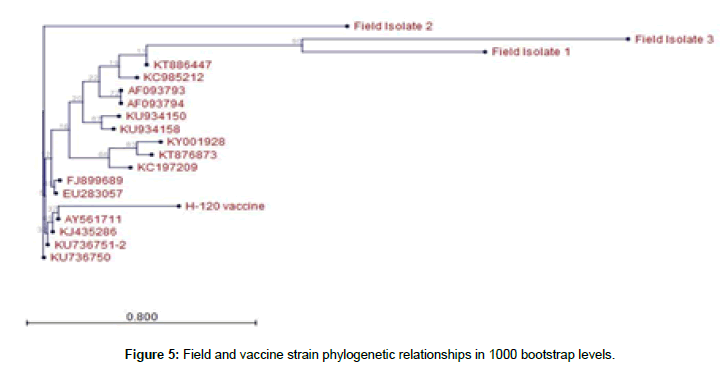
Nucleotide sequences were matched in NCBI nucleotide database (megablast) and found IBV similarity for H120 and M41 strains. Also, sequencing was performed for IBV with S1 5’ mod and CK2 primers, close similarity was found for field samples (Figure 5).
Discussion
Infectious bronchitis was first observed in the United States in 1931. Since then, disease have been detected in broiler, layer and parent flocks in worldwide. Now, studies about the disease are continuing [1,7].
There are a great number of methods for acute IBV diagnosis. Preferred diagnosis methods are virus neutralization (VN), immune fluorescent assay (IFA), immune peroxidase assay (IPA) and reverse transcriptase polymerase chain reaction (RT-PCR).
Virus neutralization test (VN) is an expensive and time consuming procedure. Therefore, it is used rarely in routine [11]. Immune fluorescent assay (IFA) and immune peroxidase assay (IPA) that introduce viral antigen in infected tissue section are used in diagnosis of IBV, but sensitivity and specify are less then molecular assays. IFA and IPA are not efficient to differentiate to serotypes and strains of IBV. Enzyme linked immune sorbent assay (ELISA) is faster and simpler then VN, but it does not detect all strains or types of IBV. Also, hemagglutination inhibition (HI) test is reliable but it can detect some strains of IBV [10].
Molecular assays are commonly used because they provide highly specific and sensitive results in a timely manner. Real time RT-PCR is more sensitive and specific than other molecular assays, because this assay uses specific primers and probe. Also, quantification of virus or its genetic material can be detected [10].
Callison et al. isolated virus from trachea using by tracheal swab. Also, it was reported that IBV can be detected following infection with the real-time RT-PCR test described herein is 21 days post-infection [10]. In most of the studies, virus was multiplied in embryonated chicken eggs before the real time PCR process [10,16].
In this study, isolation of virus was performed from trachea using by tracheal swab and then virus was multiplied in embryonated chicken eggs. Embryonic defects derived from IBV were observed. Also, the required minimum amount of RNA was guaranteed indirectly to increase the number of virus particles.
Bochkov et al. [17] found IBV positivity using by RT nested PCR. This study is related to etiology of IBV in Russia. In the study, they amplify conserved regions in 3’UTR. They found that previously known strains of IBV have similarity rate of 47 -75 % with new variant isolates in Russia and similarity among variant strains are 65 – 99 %.
Callison et al. [18] developed a new real time RT PCR procedure to recognise 5 different IBV serotype using by specific chimeric oligonucleotides. This method differentiated Massachusetts, Connecticut, Arkansas and Delaware/Georgia 98 serotypes. But they did not differentiate variant strains of IBV.
Alvarado et al. [19] used real-time RT-PCR to detect Massachussets and Arkansas serotypes of IBV. In this study, they detected presence of Massachussets and Arkansas serotypes from tracheal and secal tonsil samples using by real time RT-PCR. These serotypes were originated to two different commercial vaccines.
Callison et al. [10] investigated conserved regions in 5’UTR of IBV using by real time RT-PCR based on Taqman® hydrolyze probe. In the study, IBV and TCoV (turkey coronavirus) was detected by specific primers and probe. Other pathogens tested were not detected. In clinical experiments, although they detected 27 % IBV positivity by virus isolation, they detected 79 % IBV positivity using by TaqMan based real time RT-PCR.
In this study, conserved regions of the 5’UTR of IBV were investigated by hydrolysis probe-based Taqman real-time RT-PCR. Real-time RT-PCR protocol by Callison et al. was optimized for ROCHE Light cycler real-time PCR device [10]. In this protocol, positive results were obtained in 33 samples of 120 (27.5 %). Because primers and probe is specified to IBV, samples were considered as positive for IBV.
In detection of phylogenetic relationship of IBV and presenting of strain existence, S1 gene sequencing is most useful method [11].
Liu et al. performed amplification of 1680 nucleotide at the beginning of S1 region using by five different primers in China and introduced a phylogenetic map. In this study, Liu et al. found that similarity less than 81.1 % between vaccine strain and field strain according to S1 gene nucleotide sequence [20].
Lee et al. amplified hyper variable regions in S1 of IBV by developing general primer sets. They also carried out to molecular typing of 7 field isolates by sequencing 450 bp regions of HVR1 and HVR2. In this method, the short length (400 -450 bp) products are provided for obtaining an advantage for sequencing [21].
OIE advices amplifying of 700 bp regions of S1 containing hyper variable regions by using S15’ mod and CK2 primers for genotyping studies [14].
In this study, both C2U and Ag0723 primers reported by Lee et al. [20] were used and S1 oligo 5 and CK2 primers issued by OIE 2008 terrestrial manual and Gelb et al. [12] in order to amplify and sequence for S1 gene. After the amplification and electrophoresis of S1 gene region by C2U and Ag0723 primers, 400 – 450 bp bands belong to M41 and H120 control strains were obtained. Any bands belong to field strains and 4/91 vaccinal strain were observed. Therefore, it was considered that these field strains can be different form previously known strains and vaccinal strains which contains of both H120 and M41. Sequences of both IBV H120 and M41 strains were compared to Gene bank database and confirmed that these controls belong to IBV strains. Thereafter, 700 bp regions of S1 gene were amplified by using S15 mod and CK2 primers which advised by OIE et al. [14]. Bands between 700 and 800 bp related to 11 field samples were obtained. Some of the sequencing results related to these bands was matched to previously known IBV strains at the rate of 72 – 86 %.
In this study positive results were obtained for 33 (27.5 %) of field samples by Real Time RT-PCR. Positivity rate of 27.5 % is significant and indicates that IBV is a very contagious disease. While IBV is spreading to flocks, mutations of virus are continuing. Therefore, virus may lead to escape from host immune system [22].
Also this study have shown that direct sampling from trachea is enough for virus detection. Positive signals were obtained by real time RT-PCR when we extracted viral RNA directly from viral transport medium. But we obtained higher Ct value at direct sampling from VTM than inoculation of samples into embryonating eggs. Higher Ct value means that virus amount is less in the samples. Therefore, virus amount in the VTM is less than samples passaged into SPF eggs [23].
Sequencing of hyper variable regions of S1 gene was carried out for H120 and M41 control strains. For field samples, positive bands were not obtained after amplification with C2U and Ag0723 primers and electrophoresis. H120 and Massachusetts strains should have been given positive bands according to Lee et al. because vaccine strain used for these flocks are originated from H120 [20]. Therefore it was considered that strain that caused disease is different form vaccinal strain and Massachusetts strain [20]. Also it was considered that these virus may be a new variant. All results from sequencing of field samples is similar.
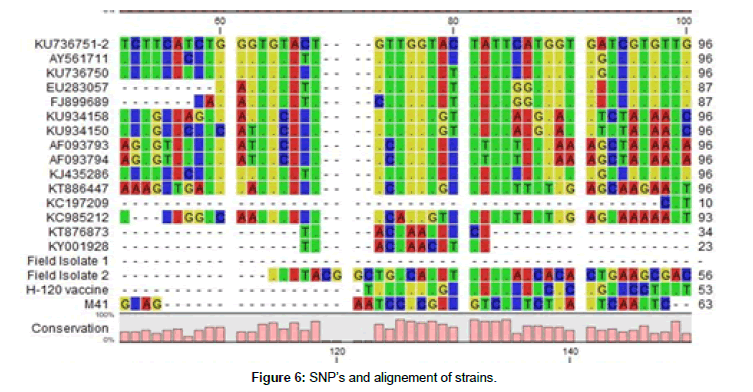
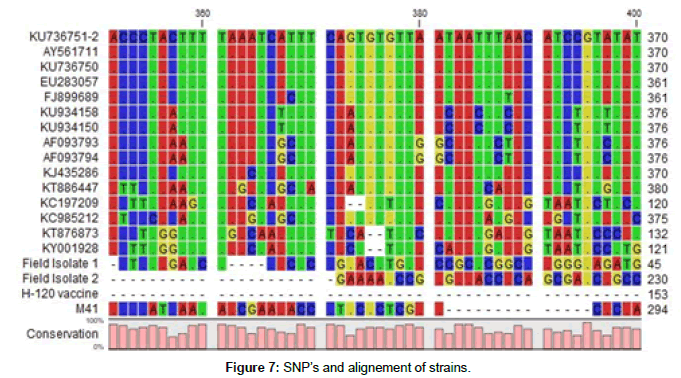
Thereafter, 700 bp bands were obtained with S1 oligo 5 and CK2 after amplification and electrophoresis, some sequecing results were matched to some variant IBV strains in gen banks database at the rate of 72-86 %. These results support that we are faced with variant IBV strains. Also we observed a lot of SNP mutations in IBV S1 gene alignement (Figures 6 and 7).
All these results indicate that IBV strains which was detected positive in this study can be variant strains. Therefore, it was considered to analyse these strains by full S1 gene sequencing with different primer sets. The identification of these field strains was suggested.
Our study has shown that using of Massachusettes seroype vaccines only does not provide good protection. Vaccination programs that is used of these vaccines only does not enough to protect flocks against variant strains and other heterolog field strains. It is adviced that after the vaccination with Massachusetts serotypes, vaccines in variant serotypes in the fields should be applied as a booster.
Conflict of Interests Statement
The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement
The research and the article were financed with the funds of Istanbul University (Project No. 3375).
Animal Rights Statement
The authors declare that the experiments on animals were conducted in accordance with local Ethical Committee laws and regulations as regards care and use of laboratory animals.
References
- Cavanagh D (2007) Coronavirus avian infectious bronchitis virus Review Article Vet Res 38: 281-297.
- Cavanagh D, Naqi SA (2003) Infectious bronchitis. (11th Edtn).
- Kannan Ganapathy, Christopher Ball, Anne Forrester (2015) Genotypes of infectious bronchitis viruses circulating in the middle east between 2009 and 2014, Virus Research 210: 198-204.
- Sen A, Sonmez G, Caner V, Ozyigit MÖ (2002) Infeksiyoz Bronsitis Virusunun Trakeal Organ Kulturlerinde immunoperoksidaz Teknigi ile Saptanmasi. Turk J Vet Anim Sci 26: 1381-1388.
- Cavanagh D (2005) Coronaviruses in poultry and other birds. Avian Pathology 34: 439-448.
- Gang X, Xiao-yu L, YeZ, Yang C, Jing Z et al (2016) Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virol J 13: 40-49.
- Butcher GD, David PS, Richard DM (2002) Infectious Bronchitis Virus: Classical and Variant Strains.
- Cavanagh D, Mawditt K, Britton P, Naylor CJ (1999) Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions. Avian Pathol 28: 593-605.
- Sahar Abd El Rahman, Hoffmann M, Lueschow D, Eladl A, Hafez HM (2015) Isolation and Characterization of New Variant Strains of Infectious Bronchitis Virus in Northern Egypt. Adv Anim Vet Sci 3: 362-371.
- Callison SA, Deborah AH, Tye OB, Brenda FS, Robert R, et al. (2006) Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J Virol Methods 138: 60-65.
- De Wit JJ (2000) Detection of infectious bronchitis virus Avian Pathology 29: 71- 93.
- Farsang A, Ros C, Lena HMR, Baule Claudia, Soós, T (2002) Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement, Avian Pathology 31: 229-236.
- Gelb JJ, Jackwood MW (2008) Infectious bronchitis, a laboratory manual for the isolation, identification and characterization of avian pathogens. (5th Edtn), American Association of Avian Pathologists, Florida, US.
- Guy JS (2005) Isolation and Propagation of Coronaviruses in Embryonated Eggs, SARS-and Other Coronaviruses Laboratory Protocols.
- OIE (2008) Infectious bronchitis 2: 443-455.
- Lee Chang-Won, Jackwood MW (2000) Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch Virol 145: 2135–2148.
- Bochkov YA, Batchenko GV, Lidiya OS, Borisov AV, Drygin VV (2006) Molecular epizootiology of avian infectious bronchitis in Russi., Avian Pathol 35: 379-393.
- Callison SA, Hilt DA, Jackwood MW (2005) Rapid differentiation of avian infectious bronchitis virus isolates by sample to residual ratio quantitation using real-time reverse transcriptase-polymerase chain reaction. J Virol Methods 124: 183–190.
- Alvarado IR, Villegas P, El-Attrache J, Jackwood MW (2006) Detection of massachusetts and arkansas serotypes of infectious bronchitis virus in broilers. Avian Dis 50: 292- 297.
- Liu Shengwang, Chen Jianfei, Han Zongxi, Zhang Qinxia, Shao Yuhao, et al. (2006) Infectious bronchitis virus: S1 gene characteristics of vaccines used in china and efficacy of vaccination against heterologous strains from china. Avian Pathol 35: 394-399.
- Lee CW, Deborah AH, Mark WJ (2003) Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene. J Vet Diagn Invest 15: 344-348.
- Shi Peng, Yu Li, Fu Yun-xin, Huang Jing-Fei, Zhang Ke-Qin, et al. (2006) Evolutionary implications of avian infectious bronchitis virus (AIBV) analysis. Cell Research 16: 323-327.
- Jackwood MW, Hilt DA, Callison SA (2003) Detection of infectious bronchitis virus by real-time reverse transcriptase–polymerase chain reaction and identification of a quasispecies in the beaudette strain. Avıan Dis 47: 718-724.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi