Research Article, J Liver Disease Transplant Vol: 6 Issue: 4
Microrna-150 and Microrna-101 As Diagnostic Markers for Hepatitis B Virus Related Hepatocellular Carcinoma in Egypt
Safaa M1*, Marwa M1 and Noha E2
1Department of Clinical Pathology, Zagazig University, Egypt
2Department of Tropical Medicine, Zagazig University, Egypt
*Corresponding Author : Saffaa Mahmoud El alawi
Department of Clinical Pathology, Faculty of Medicine, Zagazig University, Egypt
Tel: +201221551724
E-mail: saffaa_m@yahoo.com
Received: October 05, 2017 Accepted: October 17, 2017 Published: October 24, 2017
Citation: Safaa M, Marwa M, Noha E (2017) Microrna-150 and Microrna-101 As Diagnostic Markers for Hepatitis B Virus Related Hepatocellular Carcinoma in Egypt. J Liver Disease Transplant 6:4. doi: 10.4172/2325-9612.1000156
Abstract
Background: Hepatocellular carcinoma (HCC) is an increasing difficult problem in Egypt. MicroRNAs are defined as small RNAs that are non-coding RNAs which regulate gene expression after transcription. Circulating microRNAs are promising biological noninvasive markers for cancer.
Aim: The aim of this study was to evaluate miRNAs in Egyptian HBV related Hepatocellular carcinoma and their relation to clinical parameters.
Methods: MiR-150 and miR-101 relative expression were evaluated by real-time quantitative RT-PCR in 70 serum samples (20 from controls, 25 from chronic hepatitis B (CHB) patients and 25 samples from HBV related HCC patients), the diagnosis of HCC was based on typical imaging finding by dynamic computed tomography.
Results: Our results revealed that the serum relative expression levels of circulating miR-150 were lower and miR-101 were higher in patients with HCC when compared with healthy controls and CHB without HCC patients (p<0.0001). By using Receiver operating characteristic curve analyses it was suggested that miR-150 in serum had significant diagnostic value for HCC in chronic HBV patients with 75 % sensitivity and 90 % specificity. Also miR-101 in serum had significant diagnostic value for HCC in chronic HBV patients with 90 % sensitivity and 90 % specificity.
Conclusion: In conclusion, both miR-150 and miR-101 relative expression in serum in patients with HBV-related HCC could be used as noninvasive biomarkers for the early detection of HBVrelated HCC patients.
Keywords: HBV related HCC; MicroRNA; Biomarker; Real time PCR
Introduction
Hepatocellular carcinoma (HCC) is one of the most common cancers in the world and the third most common cause of death from cancer [1]. In Egypt HCC was reported to happen in nearly 4.7% of patients with chronic liver disease [2]. The main causes of HCC are both hepatitis viruses B and C (HBV) which account for more than 80% of HCC cases worldwide, HBV was the main etiologic factor in the HCC development, The annual risk of HCC is 0.5% among asymptomatic HBs Ag carriers and 0.8% for those with chronic hepatitis B, on the other hand patients with HBV-cirrhosis have 1000 times higher risk of HCC development, compared to HBs Ag negative individuals [1,3].
MiRNA are non-coding small RNA that interferes with gene expression, and it was reported that there are merely 1000 of these sequences in the human genome [4]. Each miRNA regulates its target genes negatively through binding to multiple mRNA. MiRNA and mRNA binding initiates the RNA-mediated RNAi pathway, this leads to cleavage of mRNA transcripts by a miRNA-associated RNAinduced silencing complex (miRISC) [5]. Most genes that encode miRNA located in cancer-associated regions of the genome which are fragile sites, this suggests that miRNA may be used as diagnostic markers or be therapeutic targets [6].
MiRNA influence the critical cellular functions such as proliferation, differentiation, apoptosis, invasion and metastasis [7]. In tumors the miRNA expression profiles differ from their profiles in normal tissues and also differ according to the tumor type. MiRNA targets directly protein-coding genes of the cell cycle, metastasis and apoptosis in HCC [8].
Different studies reported a significant relationship between miRNA and hepatocellular carcinoma (HCC) [9]. Some of these studies detect up regulation of the expression levels of some miRNA such as miR-21, miR-221 and miR-222, the [9-11]. Other studies detect down regulation of other miRNA, such as miR-122a, miR- 145, miR-199a and miR-223, in HCC compared to normal tissues, so miRNAs can function as oncogene or tumor suppressor to promote or to prevent HCC development [12,13].
Chang et al. [14] found that miR-150 could affect levels of Myc in Myc-mediated tumor genesis [14]. Furthermore Zhang et al. [15] reported that miR-150 by targeting cMyb could inhibit CD133- positive liver cancer stem cells. These data indicates that miR-150 might function as a tumor suppressor in HCC. And this information was interesting to evaluate whether serum miR-150 could be used as a new biomarker for the detection of HCC in chronic HBV patients [15].
On the other hand Fu et al., [16] found that miR-101 relative expression in serum in patients with HBV-related HCC was increased significantly than that in the serum of control subjects. Also, HBVx protein down regulates miR-101. MiRNAs is present in body fluid as serum and plasma, they are accessible and stable in circulation, and so they are ideal non-invasive biomarkers [17].
In our study, we evaluated whether both serum miR-150 and miR-101 relative expression in patients with HBV-related HCC could serve as a noninvasive biomarker for the diagnosis of HBV-related HCC patients.
Methods
Patients
This is a case control study conducted in Clinical Pathology and Tropical Medicine Department, The study included 70 subjects; 20 apparently healthy subjects (with no abnormality in liver biochemistry, no history of liver disease and negative viral marker) as a control group, 25 chronic HBV patients with diagnosis depend on elevated serum transaminase two fold at least in two separate setting in the last 6 month with proved serological marker for HBV and 25 patients with HBV related HCC assessed by dynamic computed tomography with typical imaging, TNM staging determined by the American committee on cancer and united international consensus committee and the staging system of Barcelona Clinic Liver Cancer (BCLC) was obtained which accounts for different factors of performance status, tumor burden and hepatic function and categorizes patients into 5 stages. All groups were age and sex matched [2].
Non HBV HCC Patients were excluded from the study. Samples from HCC patients were collected from October 2015 till October 2016. All patients were subjected to history taking, clinical examination, imaging study (Triphasic computed tomography) with laboratory investigation including CBC, liver and kidney function, viral marker HBs Ag, HBV DNA, HCV Ab, AFP, RNA extraction from serum for miRNA 101,150. The study protocol was approved by the institutional ethics committee and written informed consent was obtained from all participants.
Measurement of relative expression of microRNA 150 and 101 by real time PCR
RNA extraction: Sample processing was performed within 4 h after withdrawn from patient and controls. All samples were centrifuged at 3400 g for 7 min at room temperature. The serum was transferred into 1.7 ml eppendorf tubes followed by further centrifugation at 12000 g for 10 min at 4°C. Then the serum was separated and stored at -80°C until RNA extraction.
Total RNA was extracted from 500 μl of serum by the use of a miRNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instruction for liquid samples.
RNA conversion to complementary DNA (cDNA): RNA was reverse transcribed using miScript II RT Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions.
Reactions consisted of 15-μl RNA, 4-μl 5x miScriptHiSpec Buffer containing nucleics mix and 1 μlmiScript Reverse Transcriptase. Reactions were incubated for 60 min at 37°C then for 5 min at 95°C. The temperature scheme was performed using Gene Amp PCR System 9700 thermocycler (Perkin Elmer, Singapore). Finally, cDNA samples were Stored at –80°C till analysis.
MicroRNA quantification by real-time quantitative RT-PCR (qPCR): SYBRGreen PCR kit protocol for a StepOne Plus system (Applied Biosystems, Foster City, CA, USA) was used. qPCR reaction with 25 μl final volume was reached by adding 2.5 μl diluted cDNA, 12.5 μl 2x QuantiTect SYBR Green PCR Master Mix, 2.5 μl 10x miScript Universal Primer, 2.5 μl 10x miScript Primer Assay and 5 μl PCR grade water into real-time PCR wells. The cycling conditions were 95°C for 15 min followed by 40 cycles (94°C for 15 sec, 55°C for 30 sec, and 70°C for 30 sec). The transcription levels of target miRs were normalized to those of miR-16 which used as internal control. The normalized quantity of the target miRNA was obtained by subtracting cycle threshold (CT) for miR-16 from the CT for the target miRNA (ΔCT sample). The same calculation was performed with controls (ΔCT control). Then ΔΔCT was calculated as the difference of these values (ΔΔCT = ΔCT sample – ΔCT control). Finally, the relative expression was expressed as fold change by 2− ΔΔCT relative to control [18].
Statistical Analysis
Statistical analysis was carried out with SPSS® statistical software version 16.0 (SPSS Inc., Chicago, IL, USA).Shapiro-Wilk test was used to verify the normality of distribution of continuous variables. Data are presented as mean ± SD or median and range for continuous variables, frequency for categorical ones. Comparisons between quantitative variables were done using Anova or Kruskal-Wallis or Mann Whitney U test when appropriate. For comparing categorical data chi-squared test was performed. Receiver operating characteristic (ROC) analysis, area under curve (AUC) and 95% CI were used to determine the optimum cutoff value for the studied diagnostic markers and their diagnostic efficiencies. p -value<0.05 was considered as significantly different.
Results
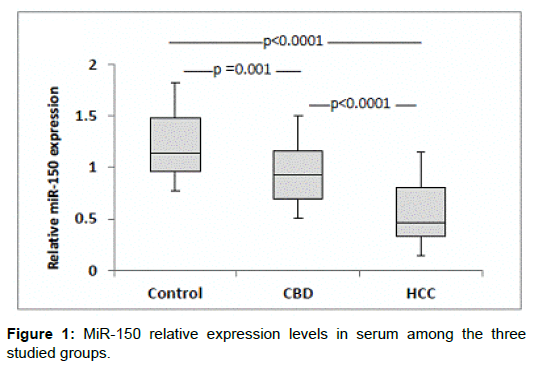
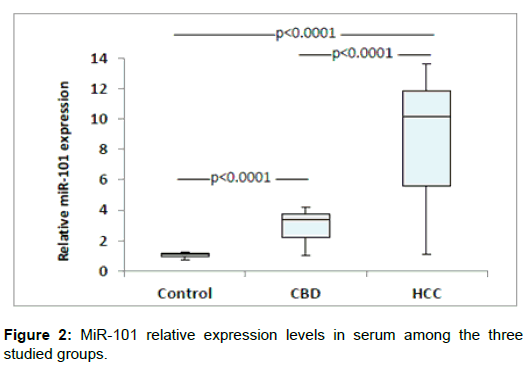
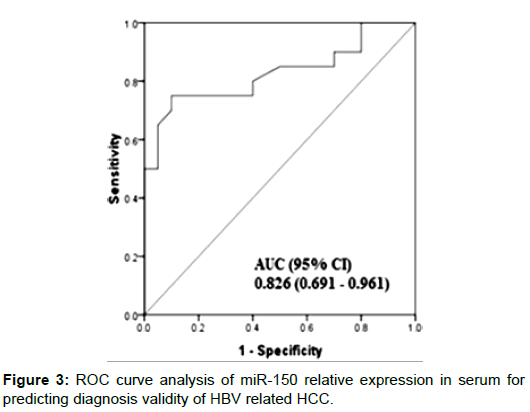
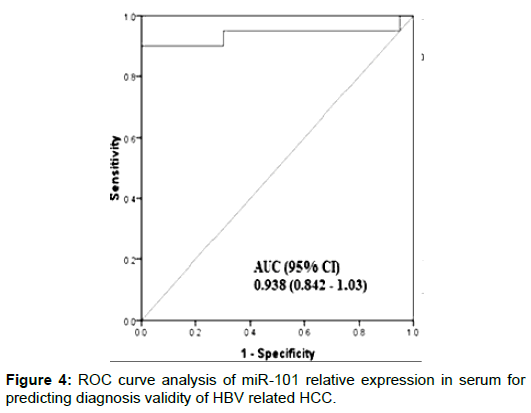
The demographic and clinical characteristics of the studied groups are described in Table 1. There were significant differences of AFP between groups (p<0.001, Kruskal-Wallis). MiR-150 relative expression levels in serum were significantly decreased in HCC patients when compared with other group (P<0.0001). Also levels were decreased significantly in CHB patients compared with controls (P = 0.001) (Figure 1). On the other hand, Serum miR-101 relative expression levels were increased significantly in HCC patients when compared with the CHB patients (P<0.0001) and healthy controls (P<0.0001). Also serum levels were significantly increased in CHB patients when compared with controls (P<0.0001) (Figure 2). Serum miR-101, 150 relative expression levels were not statistically associated with patient’s age, sex, and AFP or tumor size but were associated with child-pugh score and TNM staging (Table 2). According to miR-150 median value (0.46) and miR-101 median value (10.1) we divided the serum samples into low expression and high expression groups. The association between miR-150 and miR-101 expression and clinical parameters was analyzed (Table 2). We did not find significant association between low and high miR-150 and miR-101 expression as regard BCLC stages (Data not shown). ROC-AUC of miR-150 relative expression in diagnosis of HCC. At the cutoff value of 0.66, the sensitivity and the specificity were 75% and 90%, respectively (Figure 3). ROC-AUC of miR-101 relative expression in diagnosis of HCC. At the cutoff value of 4.15, the sensitivity and the specificity were 90% and 90%, respectively (Figure 4).
| Characteristic | HCC | CHB | Controls | p |
|---|---|---|---|---|
| (n=25) | (n=25) | (n=20) | ||
| Age | ||||
| Mean ± SD | 60.2 ± 5.8 | 58.7± 6.6 | 58.3 ± 6.2 | 0.597 |
| Sex | ||||
| Male / Female | 18/7 | 16/9 | 14/6 | 0.92 |
| AFP(ng/ml) | ||||
| Median | 600 | 18 | 2.15 | <0.001* |
| Range | 20 - 6051 | 3 - 60 | 0.8 – 13 | |
| HBsAg | ||||
| Positive | 25(100%) | 25 (100%) | 0 | |
| Negative | 0 | 0 | 20(100%) | |
| cirrhosis | ||||
| + | 21(84%) | 0 | 0 | |
| - | 4(16%) | 0 | 0 | |
| Child-Pugh | ||||
| A | 6(24%) | 0 | 0 | |
| B | 19(76%) | 0 | 0 | |
| Tumor diameter (cm) | ||||
| <5 | 15(60%) | |||
| >5 | 10(40%) | |||
| TNM staging | ||||
| I | 7 (28%) | |||
| II | 14 (56%) | |||
| III | 4 (16%) | |||
| IV` | 0 (0%) | |||
| BCLC | ||||
| A | 19 (76%) | |||
| B | 6 (24%) | |||
Table 1: Clinical characteristics and demographic data of the three studied groups.
| Parameter | miR-150 (N=25) |
p | miR-101 (N=25) |
P | ||
|---|---|---|---|---|---|---|
| Low | High | Low | High | |||
| Age n=13 n=12 n=10 n=15 | ||||||
| < 60 | 5 | 6 | 0.56 | 4 | 6 | 1.0 |
| ≥ 60 | 8 | 6 | 6 | 9 | ||
| Sex | n=14 | n=11 | n=14 | n=11 | ||
| Male | 10 | 8 | 0.94 | 11 | 7 | 0.41 |
| Female | 4 | 3 | 3 | 4 | ||
| AFP | n =15 | n =10 | n =12 | n =13 | ||
| <200 | 7 | 4 | 0.74 | 5 | 7 | 0.55 |
| ≥ 200 | 8 | 6 | 7 | 6 | ||
| Child-Pugh | n=6 | n =15 | n =6 | n =15 | ||
| A | 1 | 10 | 0.043* | 4 | 4 | 0.046* |
| B | 5 | 5 | 2 | 11 | ||
| Tumor n =12 n =13 n =12 n =13 diameter | ||||||
| <5 | 6 | 9 | 0.33 | 7 | 8 | 0.87 |
| ≥5 | 6 | 4 | 5 | 5 | ||
| TNM staging | n =9 | n =16 | n =14 | n =11 | ||
| I | 1 | 9 | 0.002* | 7 | 1 | 0.02* |
| II | 2 | 5 | 3 | 2 | ||
| III | 6 | 2 | 4 | 8 | ||
Table 2: The relation between miR-150 and miR-101 relative expression in serum and clinical parameters of HCC patients.
Discussion
Hepatocellular carcinoma (HCC) is a nightmare because it is considered the most common primary liver cancer that causes around 700,000 mortalities worldwide yearly [19]. Many of HCC cases occur in top chronic hepatitis B virus (HBV) infection. HBV was the main etiologic factor in the development of HCC. The annual risk of HCC is 0.5% among asymptomatic HBsAg carriers and 0.8% for those with chronic hepatitis B, on the other hand patients with HBV-cirrhosis has 1000 times higher risk of the development of HCC, compared to HBsAg negative individuals [20]. The challenge in HCC is that most of cases with HCC are lately diagnosed, and late stages of HCC associated with a poor prognosis and low survival rates because of lack of curative treatment options. And so early detection of HCC is required, the available screening approach of HCC in a high risk‑population is determining serum tumor markers, such as α‑fetoprotein (AFP). But, AFP is not sensitive for early small sized tumors and not specific for HCC as it can be elevated in benign conditions. Thus, new biochemical markers for the diagnosis of HCC in early stages are required [21,22].
New class of RNA molecules termed microRNAs (miRNAs) [which are small in size and have regulatory function on gene expression] that are involved in HCC development and progression are described. MiRNAs negatively regulate gene expression by interacting with the 3untranslatedregion (UTR) of protein-coding mRNA. By formation of the RNA-induced silencing factor complex, miRNAs binding generally causes translational suppression and/or degradation of the target transcript [23,24]. MiRNAs implicated in several cellular processes, including cell proliferation, differentiation, and tumorigenesis [25]. The role of miRNAs in cancer pathogenesis has been well established, as they may act as tumor suppressor genes or oncogenes [26].
Our study showed that serum miR-150 serum relative expression levels were significantly decreased in HCC patients in compare with other group, Also, It was significantly decreased in CHB patients when compared with control subjects, this was in accordance with Yu et al. [27]. This result suggests that miR-150 may serve as a tumor suppressor in the HCC development [28]. Also, low serum miR- 150 was significantly associated with child-paugh score and TNM staging, this suggests that miR-150 is associated with liver function and prognosis of HCC [29,30]. At cut off value of 0.66, the diagnostic validity of miR-150 (AUC=0.82) for HBV related HCC the sensitivity was 75% the specificity 90% respectively. This was in agreement with YU et al. [27] who showed that miR-150 diagnostic value for HBV related HCC with sensitivity 79.1% and specificity 76.5%. So serum miR-150 could be used for diagnosis and prediction of the prognosis of HBV related HCC.
In contrast, our study has showed that serum levels of miR-101 expression were significantly increased in in HCC patients when compared with both CHB patients and controls. Also, miR-101 expression levels in serum were higher with statistically significant increase in CHB patients than healthy group, This was in agreement with Fu et al. [16] Who found that serum miR-101 level increased by approximately 10 fold compared with healthy control so miR-101 may be important in HBV related HCC tumor genesis. MiR-101 may pass to the circulation by excessive secretion by HCC cancer cells, [14,31] or tissues affected by the disease [32]. The deregulated miR-101 may contributes to the activation of oncogenes and the inactivation of tumor suppressor genes in human carcinogenesis [33,34].
As regard serum miR-101 and clinical characteristic of HCC patients, it was not associated with age, Gender, AFP and tumor size however statistically significant association was detected between miR 101 and Child-pugh score and TNM staging, this was in agreement with FU et al. [16] suggesting that miR101 may be associated with liver function and might serve as prognostic marker.
Our current data, showed that at cut off value of 4.15, miR101 had a diagnostic validity (AUC=0.93) where, the sensitivity and the specificity for diagnosis of HBV related HCC were 90% and 90% respectively, This suggest that miR101 may be a potential marker for HBV related HCC.
Conclusion
In conclusion, both serum miR-150 and miR-101 expression in patients with HBV-related HCC could serve as a noninvasive biomarker for diagnosis of HCC in chronic hepatitis B patients. And it’s recommended to explore for serum microRNA panel to be used for early diagnosis of HCC.
References
- Parkin DM (2000) Global cancer statistics in the year. Lancet Oncol 2: 533-543.
- McGlynn KA, Tsao L, Hsing AW, Devesa SS, Fraumeni JF Jr (2001) International trends and patterns of primary liver cancer. Int J Cancer 94: 290-296.
- Bosch FX, Ribes J, Díaz M, Cléries R (2004) Primary liver cancer: worldwide incidence and trends. Gastroenterology 127: 5-16.
- Zamore PD, Haley B (2005) Ribo-gnome: the big world of small RNAs. Science 309: 1519-1524.
- Suzuki H, Maruyama R, Yamamoto E, Kai M (2013) Epigenetic alteration and microRNA dysregulation in cancer. Front Genet 4: 258.
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. ProcNatlAcadSci U S A 101: 2999-3004.
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646-674.
- Liu WR, Shi YH, Peng YF, Fan J (2012) Epigenetics of hepatocellular carcinoma: a new horizon. Chin Med J (Engl) 125: 2349-2360.
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, et al. (2007) MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 133: 647-658.
- Wong QW, Lung RW, Law PT, Lai PB, Chan KY, et al. (2008) MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 135: 257-269.
- Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, et al. (2008) MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology 47: 1223-1232.
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, et al. (2007) Cyclin G1 is a target of miR-122a, a microRNA frequently downregulated in human hepatocellular carcinoma. Cancer Res. 67: 6092-6099.
- Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, et al. (2006) Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 25: 2537-2545.
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, et al. (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 40: 43-50.
- Zhang J, Luo N, Luo Y, Peng Z, Zhang T, et al. (2012) MicroRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 40: 747–756.
- Fu Y, Wei X , Tang C, Li J, Liu R, et al. (2013) Circulating micro RNA-101 as a potential biomarker for hepatitis B virus‑related hepatocellular carcinoma. Oncology letters 6: 1811-1815.
- Weber JA, Baxter Dh, Zhang S, Huang DY, Huang KH, et al.( 2010) The microRNA spectrum in 12 body fluids. Clin Chem. 56: 1733-1741.
- Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402-408.
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69-90.
- Arbuthnot P, Kew M (2001) Hepatitis B virus and hepatocellular carcinoma. Int J ExpPathol. 82: 77-100.
- Oka H, Tamori A, Kuroki T, Kobayashi K,Yamamoto S (1994) Prospective study of alpha-fetoprotein in cirrhotic patients monitored for development of hepatocellular carcinoma. Hepatology 19: 61-66.
- Marrero JA, Lok AS (2004) Newer markers for hepatocellular carcinoma. Gastroenterology 127: 113-19.
- Guo H, Ingolia NT, Weissman JS, David PB, et al. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835-840.
- Gottwein E, Cullen BR (2008) Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Hostand Microbe 3: 375-387.
- Bushati N, Cohen SM (2007) MicroRNA functions. Annual Review of Cell and Developmental Biology 23: 175–205.
- Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482: 347-355.
- Yu F, LU Z, Chen B, Peihong Dong, Jianjian Zheng (2015) MicroRNA-150: a promising novel biomarker for hepatiis B virus-related hepatocellular carcinoma. Diagnostic pathology 110: 129.
- Chen X, Ba Y, Ma L, Cai X, Yin Y et al. (2008) Characterization of microRNAs in serum:a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18: 997-1006.
- Johson Pj, Melia WM, Palmer MK, Portmann B, Williams R (1981) Relationship between serum alpha-foetoprotein, cirrhosis and survival in hepatocellular carcinoma.Br J Cancer 44: 505-505.
- Farinati F, Rinaldi M, Gianni S, Naccarato R (2000) How should patients with hepatocellular carcinoma be staged? Validation of a new prognostic system. Cancer 89: 2266-2273.
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, et al. (2010) Functional delivery of viral miRNAs via exosomes. ProcNatlAcadSci USA 107: 6328‑6333.
- Skog J, Würdinger T, van Rijn S, Dimphna Meijer, Laura Gainche, et al. (2008) Glioblastomamicrovesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 10: 1470-1476.
- Mott JL (2009) MicroRNAS involved intumor suppressor and oncogene pathways: Imlications for hepatobiliary neoplasia. Hepatology 50: 630-637.
- Visone R, Petrocca F, Croce CM (2008) Micro-RNAs in gastrointestinal and liver disease. Gastroenterology 135: 1866-1869.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi