Research Article, J Virol Antivir Res Vol: 10 Issue: 4
Molecular Analysis of HCV Real Time PCR in Multan Pakistan
Muhammad Waqar Mazhar*, Javaria Mahmood, Saira Saif Ahmad Raza, Masooma Batool Shahzadi, AmnaMaqsood, Zarsha Naureen, Qandeel Zahra, Sadaf Javeed and Hira Aslam
Department of Bioinformatics and Biotechnology, Government Collage University, 38000Faisalabad, Pakistan
*Corresponding Author: Muhammad Waqar Mazhar
Department of Bioinformatics and Biotechnology, Government Collage University, 38000Faisalabad, Pakistan
Tel: +923012222861
E-mail: waqarmazhar63@gmail.com
Received: May 08, 2021 Accepted: June 01, 2021 Published: June 08, 2021
Citation: Mazhar MW, Raza A, Sikandar M, Mahmood J, Saif S, et al. (2021) Molecular Analysis of HCV Real Time PCR in Multan Pakistan. J Virol Antivir Res 10:4.
Abstract
HCV belongs to the Flaviviridae family of viruses. Have a 9.6 kb single-standard, positive-sense RNA genome. HCV has seven genotypes and 67 subtypes. HCV patient increases day by day in world. In HCV patient LFTs test values increase in men and they have some risk factor, those people who have blood transfusion in last 6 month, some mothers have HCV it transfer from parents to their mother and unprotected sex with multiple partner.
Keywords: Chronic HCV; Flaviviridae; Aspartate Aminotransferase; Alanine ; Aminotransferase
Introduction
HCV belongs to the Flaviviridae family of viruses [1] Have a 9.6 kb single-standard, positive- sense RNA genome with a length of about 10,000 bases. Chronic liver disease, cirrhosis, and hepatocellular carcinoma are all caused by it. [2,3] HCV is endemic in Pakistan, with approximately 3% of the population chronically infected and an estimated 170 million people worldwide infected with the virus [4]. [5] HCV has seven genotypes and 67 subtypes based on homology. In Iraqi patients, subtype 1a (28 percent), 4e (24 percent), 1b (18 percent), 4a (14 percent), 4b (12 percent), and 4e (4 percent) were the most common HCV subtypes. [6] Recent data suggest that, while genotype 3a may still be the most common HCV subtype in Pakistan, the epidemiological pattern and relative frequency distribution of various genotypes are changing significantly [7]. These studies have found an increase in genotype 2a, especially in the north western province of Khyber Pakhtunkhwa and the Sindh provenance [8]
HCV is primarily transmitted through the parental pattern, but 90% of intravenous drug users are at high risk. Interferon and ribavirin are regular therapies for chronic HCV and have a 38- 43 per cent prolonged virological response [9]. In most clinical laboratories worldwide, the only clinical diagnosis that is not valuable is the anti-HCV ELISA test. Several immunological methods, as well as enzyme-linked immune absorptive agents, were used in this study. [10]
Aspartate Aminotransferase (AST) and Alanine Aminotransferase (ALT) levels indicate active liver inflammation and tissue damage during chronic hepatitis C virus (HCV) infection, while Albumin reflects synthetic liver function and nutritional status.[11] ALT, also known as serum glutamic pyruvic transaminases, low level of ALT usually found in the blood. If the liver is damaged or diseased, it releases substances into the bloodstream, indicating liver disease. There are various liver disorders, including alcoholic liver disease, acetaminophen hepatotoxicity, acute viral hepatitis, and fatty liver infiltration, [12]. Platelets are also counted in all patients. [13] In human hepatocytes, NK cells play a role in suppressing HCV infection, and replication ALT, also known as serum glutamic pyruvic transaminases, low levels of ALT are usually found in the blood.[14]
Methodology
Patients
125 samples was collected from Multan district of Punjab from 2022 to January 2017. Patient information including name, age and sex was recorded. All the patients selected for the study were positive for HCV by rapid screening and Anti HCV by Elisa. 5ml venous blood was collected from each donor by using disposable syringes. Sample is transferred to vials containing anticoagulant, centrifuged and plasma was separated for further analysis.
All tested positive for HCV RNA and negative for human immunodeficiency virus. Other causes of chronic hepatitis were excluded by appropriate serological testing and liver histology. The HCV genotype was determined by sequence analysis using the HCV real time PCR.
Sera for HCV RNA Analysis
A total of 125 serum samples were obtained from Multan and khanewal region. Each of the specimens was frozen to −80°C within 2 h of collection. Each aliquot was used for HCV RNA quantitation by an Apllied bio system instrument, with a detection limit of 18.354 IU/ml.
The HCV RNA viral load was measured at the baseline of cycle. The lower limit of detection is
18.354 IU/ml, with ≥95% probability, using 1 ml of serum.
Applied biosystem Real Time HCV Assay
The Real Time HCV assay (Apllied bio system instrument) was performed at Lahore PCR lab. According to the manufacturer’s specifications. Total nucleic acids were extracted from 0.5 ml serum or plasma by magnetic microparticle technology, using a m1000 automated sample preparation system. The internal control (IC), derived from the hydroxypyruvate reductase gene of the pumpkin plant, Cucurbita pepo, was introduced as armored RNA into the sample lysis buffer. The IC was processed simultaneously with each sample. RNA was captured by magnetic microparticles, washed to remove unbound sample components, and eluted. A second step consisted of adding master mix (HCV oligonucleotide reagent including primers and probes, thermostable rTth polymerase enzyme, and activation reagent) to extract nucleic acid samples into 96-well optical reaction plates. Plates were sealed and placed on an Applied biosystem instrument.) for reverse transcription, PCR amplification, and detection/quantitation.
The HCV probe is a short linear oligonucleotide labeled with a fluorophore at the 5′ end and a quencher at the 3′ end. The PCR primers and probe target conserved regions of the 5′ untranslated region of the HCV genome. In the absence of an HCV target, the fluorescence is quenched. In the presence of the HCV target sequence, the HCV probe preferentially hybridizes to the target sequence, allowing fluorescence detection. A second set of primers and a probe labeled with a different fluorophore are used for amplification and detection of the internal control. The IC probe is a single-stranded DNA oligonucleotide with a fluorophore at the 5′ end and a quencher at the 3′ end. IC probe fluorescence is quenched in the absence of IC target sequences. In the presence of IC target sequences, IC probe hybridization to complementary sequences separates the fluorophore and quencher and allows fluorescence emission and detection. The noncompetitive internal control is detected at all HCV levels.
The HCV- and IC-specific probes are each labeled with a different fluorophore, thus allowing for simultaneous detection of both amplified products at each cycle. The homogeneous format and sealed PCR tray eliminate contamination by amplified products.
The assay meets the Second WHO International Standard for HCV RNA. The lower limit of detection is 18.354 IU/ml, with ≥95% probability. The dynamic range of the assay extends from 12 to 100,000,000 IU/ml. The applied bio system Real Time HCV assay detects and quantitates genotypes 1 to 6.
LFT’s perform from plasma that isolated from whole blood after centrifugation. Added 800ul R1 solution in the test tube. Added 200ul R2 solution in R1solution test tube and kept at R/T for 5 minutes. After this added 100ul plasma in test tube and run on Mindray 2600DR.
Sensitivity of RT-PCR
The analytical sensitivity of the real-time RTPCR was determined with using dilution series of the HCV 5’-NCR RNA transcripts containing 0, 102, 10 3, 10 4, 10 5, 10 6 and 107 molecules and tested five times. s). The lower limit of detection is 18.354 IU/ml, with 95% probability. The dynamic range of the assay extends from 12 to 100,000,000 IU/ml. The Applied Biosystems (StepOne™ and StepOnePlus™ Real-Time PCR Systems) assay detects and quantitates genotypes 1 to 6. The sensitivity of HCV REAL TIME PCR is 18.354 IU/ml.
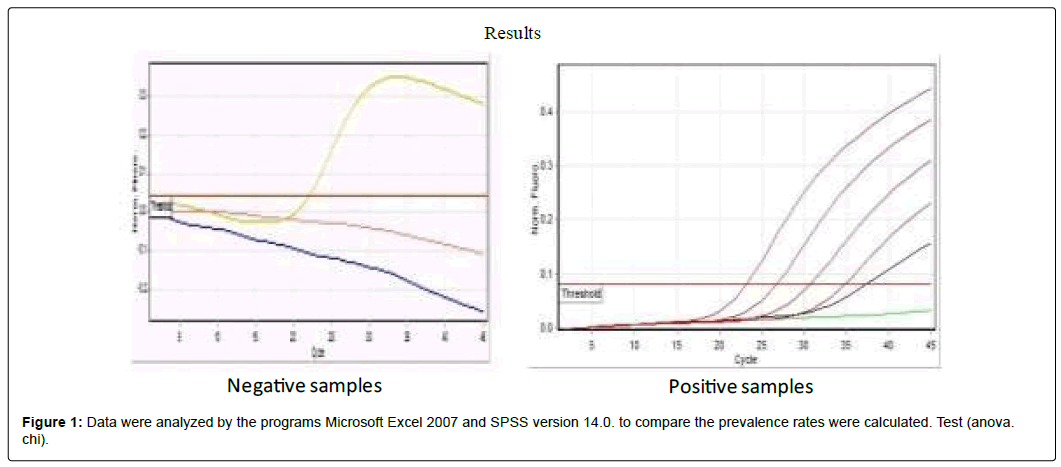
Data were analyzed by the programs Microsoft Excel 2007 and SPSS version 14.0. to compare the prevalence rates were calculated. Test (anova. chi). (Figure 1)
Typical amplification plot of HCV real time PCR. The graph of the increment of fluorescence reporter signal versus cycle number during PCR shows three stages baseline, exponential phase, and plateau. The CT value is calculated by determining the point at which the fluorescence exceeds an arbitrary threshold limit (usually 10 times the standard deviation of the baseline)
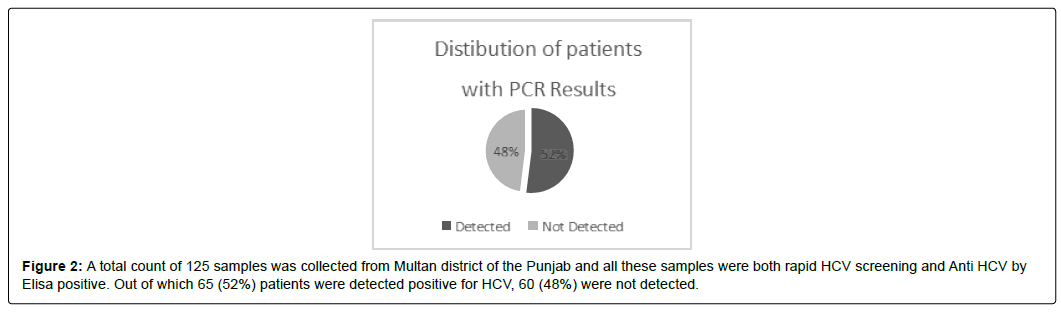
A total count of 125 samples was collected from Multan district of the Punjab and all these samples were both rapid HCV screening and Anti HCV by Elisa positive. Out of which 65 (52%) patients were detected positive for HCV, 60 (48%) were not detected. (Figure 2)
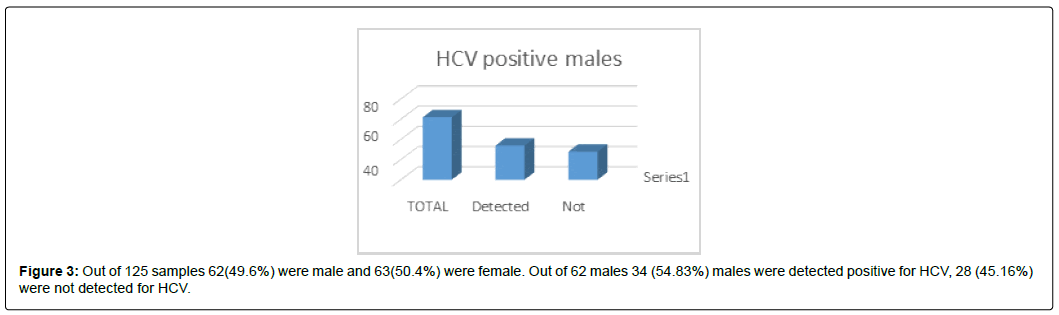
Out of 125 samples 62(49.6%) were male and 63(50.4%) were female. Out of 62 males 34 (54.83%) males were detected positive for HCV, 28 (45.16%) were not detected for HCV. (Figure 3)
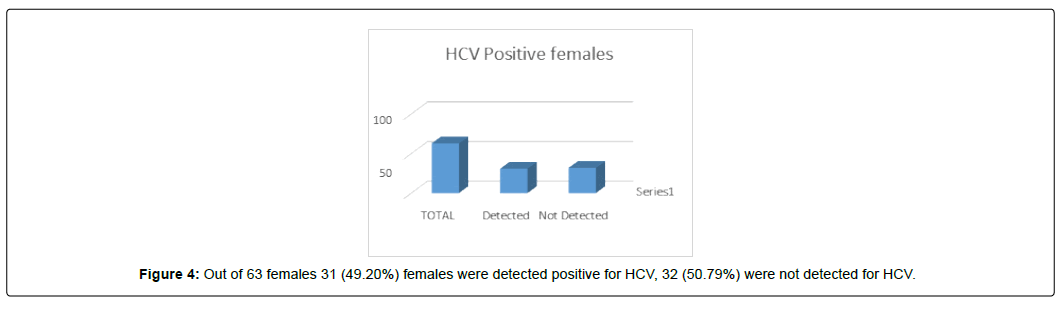
Out of 63 females 31 (49.20%) females were detected positive for HCV, 32 (50.79%) were not detected for HCV. (Figure 4)
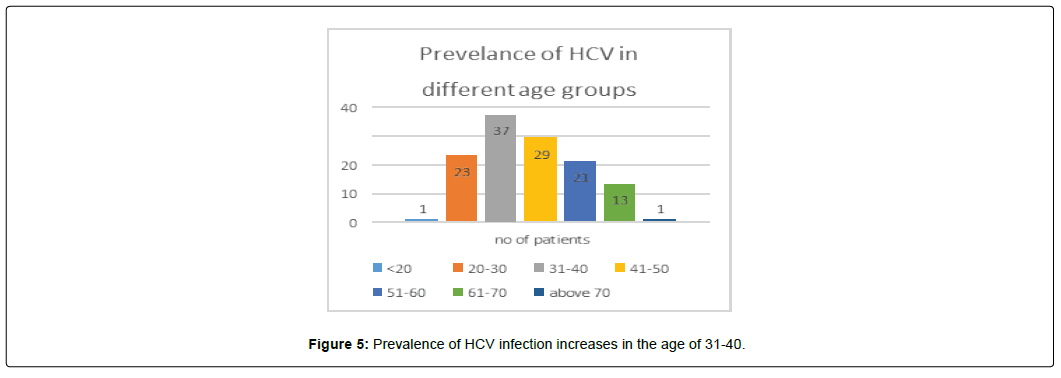
Hepatitis C 1 (0.8%) samples was from age group of <20 age that was not detected, 23(18.4%) were from 20-30 age out of which 10 were detected, 13 were not detected and only, 37 (29.6%) were from 31-40 age out of which 21 were detected positive for HCV, 16 were not detected, 29 (23.2%) samples were from 41-50 age out of which 16 were detected positive for HCV, 13 were not detected, 21(16.8%) were from 51-60 age out of which 11 were detected positive for HCV, 10 were not detected, 13(10.4%) were from 61-70 age out of which 7 were detected positive for HCV, 6 were not detected, only 1 (0.8%) was above 70 years that was also not detected. Prevalence of HCV infection increases in the age of 31-40. (Figure 5).
References
- Chandriani S (2013) Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proceedings of the National Academy of Sciences. 110(15): E1407-1415
- Moradpour D, Penin F, Rice CM (2007) Replication of hepatitis C virus. Nature Reviews Microbiology 5(6): 453 -463.
- Shepard CW, Finelli L, Alter MJ (2005) Global epidemiology of hepatitis C virus infection. The Lancet infectious diseases 5(9): 558-567.
- Mohsen A, Trent HCV Group (2001) The epidemiology of hepatitis C in a UK health regional population of 5.12 million. Gut 48(5): 707-713.
- Lavanchy D (2011) Evolving epidemiology of hepatitis C virus. Clinical Microbiology and Infection 17(2): 107-115.
- Smith DB (2014) N Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59(1): 318-327.
- Khan N (2014) Geographic distribution of hepatitis C virus genotypes in pakistan. Hepatitis monthly 14(10): e20299.
- Nazir N (2017) Prevalence of hepatitis-C virus genotypes and potential transmission risks in Malakand Khyber Pakhtunkhwa, Pakistan. Virology journal 14(1): 160.
- Munir S (2010) Hepatitis C treatment: current and future perspectives. Virology journal 7(1): 296.
- Ahmad N (2007) An evidence of high prevalence of Hepatitis C virus in Faisalabad, Pakistan.Saudi medical journal 28(3): 390-395.
- Kostadinova L (2018) Soluble Markers of Immune Activation Differentially Normalize and Selectively Associate with Improvement in AST, ALT, Albumin, and Transient Elastography During IFN-Free HCV Therapy. Pathogens and Immunity 3(1): 149-163.
- Sheth SG (1998) AST/ALT ratio predicts cirrhosis in patients with chronic hepatitis C virus infection. The American journal of gastroenterology 93(1): 44-48
- Pohl A (2001) Serum aminotransferase levels and platelet counts as predictors of degree of fibrosis in chronic hepatitis C virus infection. The American journal of gastroenterology 96(11): 3142-3146.
- Wang SH (2008)Natural killer cells suppress full cycle HCV infection of human hepatocytes. Journal of viral hepatitis 15(12): 855-864.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi