Commentary, J Pulm Med Vol: 5 Issue: 4
Molecular Dynamics at the Checkpoint
Bernhard Roither1, Chris Oostenbrink2, Georg Pfeiler3, Heinz Koelbl3 and Wolfgang Schreiner1*
1Department of Biosimulation and Bioinformatics, Medical University of Vienna, Austria
2Department of Molecular Modeling and Simulation, University of Natural Resources and Life Science, Austria
3Department of Obstetrics and Gynecology, Medical University of Vienna, Austria
*Corresponding Author: Wolfgang Schreiner Department of Biosimulation and Bioinformatics, Medical University of Vienna, Austria Tel: +431 40400 66790 E-mail: wolfgang.schreiner@meduniwien.ac.at
Received: August 16, 2021; Accepted: August 30, 2021; Published: September 06, 2021
Citation: B Roither, Ch Oostenbrink, G Pfeiler, H Koelbl, W Schreiner (2021) Molecular Dynamics at the Checkpoint, J Pulm Med 5:4
Abstract
Computational molecular dynamics and by big data analysis disclose molecular movement patterns relevant for drug development and function. Atoms in the contact zones of the PD-1 receptor move differently depending on the binding partner: the natural ligand, PD-L1, or the checkpoint inhibitors nivolumab and pembrolizumab, respectively. Computational analysis was performed by an interdisciplinary team from MedUni Vienna and the University of Natural Resources, Vienna. The natural ligand, PD-L1, not only binds but also activates PD-1, thereby inducing T-cell apoptosis. Cancer cells may, by expressing PD-L1, halt natural immune attack and thus survive. Checkpoint inhibitors are designed to just bind to PD-1, but without activating it. This functional difference is reflected by molecular movements analyzed in the papers by Roither being outlined here.
Keywords: Computational molecular dynamics
The Method of Molecular Dynamics
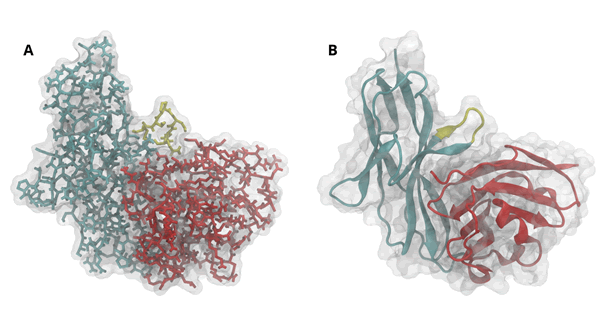
Molecular dynamics is a computational technique to compute atomic movements similar to a planetary system – but for thousands of atoms rather than nine planets. Atomic positions of all molecules (PD-1, PD-L1, nivolumab, pembrolizumab) are taken from the Protein Data Bank (PDP) [1-4] (Figure 1). Given positions, distances and forces between atoms are computed [5]. Atomic velocities are assigned randomly, according to physiological temperature. The‚ flight trajectories are then computed along a very short interval of time (2 × 10-15 sec), yielding the next configuration, i.e. new positions, distances, forces and velocities. The next step of simulation may follow [6]. The system can be watched just like in a movie. At intervals, positions are stored and represent a 'trajectory' for subsequent evaluation.
The software GROMACS [7,8] was run on the supercomputer at the Vienna Scientific Cluster (VSC) and took 5 weeks to compute the system‘s development during a real time of 200 nanoseconds.
Refined Big Data Analysis
Advanced analysis is mandatory to extract anything meaningful from the vast amount of data. For evaluation, the software Visual Molecular Dynamics (VMD) [9] and programs coded in MATLAB [10] were used.
Similarity and Clusters
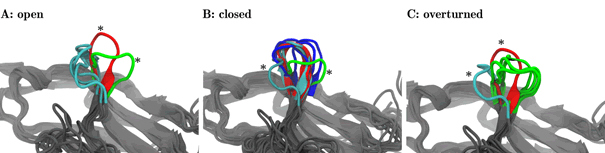
In a first step, configurations are compared and considered similar if their multidimensional distances [11] are small. Multidimensional distance may be defined across a whole molecule or with respect to some special part, such as an alpha helix, a beta sheet or a loop. Similar configurations may be grouped into clusters [12]. In our system, the CC‘- loop of PD-1 (Figure 1) proved a most relevant region for clustering.
Clusters Relate to Function
Certain clusters regarding the configuration of the CC‘-loop occur with both, the natural ligand as well as the drugs as binding partner. Other clusters occur with specific partners only. For example, one specific configuration of the CC‘-loop is only seen with Pembrolizunab as ligand it is 'specific'for this drug. Conversely, with the natural ligand, the very same loop toggles between two configurations (clusters), (Figure 2). This may represent the molecular reason why the natural ligand is able to trigger PD-1 while the drug does not it just binds and blocks the checkpoint [13-15].
Conclusion
Molecular dynamics simulation offers detailed insight into basic mechanisms of molecular medicine and is a conerstone of in silico drug design. It reveals atomnistic explanations that cannot be povided experimentally, regarding biololecules and also drugs. Nivolumab and Pembrolizumab have already been evaluated regarding lung cancer. Modified drugs may be simulated to estimate their therapeutic potential prior to wet lab or even clinical studies. While the described article focuses on PD-1, other chekpoints and corresponding inhibitors may be tackled as well using the very same techniques opening a wide range of applications of Computational Molecular Medicine.
References
- Quatrini L, Mariotti FR, Munari E, Tumino N, Vacca P, et al, (2020) The immune checkpoint pd-1 in natural killer cells: Expression, function and targeting in tumour immunotherapy. Cancers (Basel) 12(11).
- Roither B, Oostenbrink C, Pfeiler G, Koelbl H, Schreiner W (2020) Pembrolizumab induces an unexpected conformational change in the CC'-loop of PD-1. Cancers (Basel), 13(1).
- Roither B, Oostenbrink C, Schreiner W (2020) Molecular dynamics of the immune checkpoint Programmed Cell Death Protein I, PD-1: Conformational changes of the BC-loop upon binding of the ligand PD-L1 and the monoclonal antibody nivolumab. BMC Bioinformatics 21(17).
- Burley SK (2013) PDB40:The protein data bank celebrates its 40th birthday. Biopolymers 99(3):165-169.
- Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem, 25(13):1656-1676.
- Hansson T, Oostenbrink C, Van Gunsteren WF (2002) Molecular dynamics simulations. Curr Opin Struct Biol, 12(2):190-196.
- Abraham MJ, Murtola T, Schulz R, Pall S, Smith JC, et al (2015) Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2:19-25.
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, et al (2013) GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29(7):845-854.
- Humphrey W, Dalke A, Schulten K (1996) VMD: Visual molecular dynamics. J Mol Graph 14(1):33-38.
- Martinez WL, Martinez AR, Solka JL (2011) Exploratory data analysis with MATLABR, second Edition edn. London: CRC-Press
- Schreiner W, Karch R, Knapp B, Ilieva N (2012) Relaxation Estimation of RMSD in Molecular Dynamics Immuno-Simulations. Comput Math Methods Med: 173521.
- Daura X, van Gunsteren WF, Mark AE (1999) Folding Unfolding Thermodynamics of a ß-Heptapeptide From Equilibrium Simulations. Proteins, 34(3):269-280.
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, et al (2018) Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med , 378(22):2078-2092.
- Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, et al (2015) Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med, 372(21):2018-2028.
- Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, et al (2015) Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med, 373(17):1627-1639.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi