Research Article, J Diagn Tech Biomed Anal Vol: 12 Issue: 1
Sequencing the SARS-CoV-2 Genome from Stool Samples of Post-Acute Cases Implicates a Novel Mutation Associated with Reduced Antibody Neutralization
Nataliya Panova1, Nina P Allan1, Noelle C Rubas1, Rosa H Lee1, Braden P Kunihiro1, Lesley Umeda1, Rafael Peres1, Ruben Juarez2 and Alika K Maunakea1*
1Department of Anatomy, Biochemistry and Physiology, John A. Burns School of Medicine, University of Hawaii, Honolulu, HI 96813, USA
2Department of Economics and UHERO, University of Hawaii, Honolulu, HI 96822, USA
*Corresponding Author: Alika K Maunakea
Department of Anatomy,
Biochemistry and Physiology
John A. Burns School of Medicine,
University of
Hawaii
Honolulu, HI 96813
USA
E-mail: amaunake@hawaii.edu
Received date: 04 November, 2022, Manuscript No. JDTBA-22-79002; Editor assigned date: 07 November, 2022, PreQC No. JDTBA-22-79002 (PQ); Reviewed date: 21 November, 2022, QC No. JDTBA-22-79002; Revised date: 04 January, 2023, Manuscript No. JDTBA-22-79002 (R); Published date: 11 January, 2023, DOI: 10.4172/2469-5653.1000267
Citation: Panova N, Allan NP, Rubas NC, Lee RH, Kunihiro BP, et al. (2023) Sequencing the SARS-CoV-2 Genome from Stool Samples of Post-Acute Cases Implicates a Novel Mutation Associated with Reduced Antibody Neutralization. J Diagn Tech Biomed Anal 12:1
Abstract
Objective: Whole genome SARS-CoV-2 sequencing tools are crucial for tracking the COVID-19 pandemic. However, current techniques require sampling of actively infectious patients following COVID-19 testing to recover enough SARS-CoV-2 RNA from the nasopharyngeal passage, which rapidly clears during the first few weeks of infection. A prospective assessment of the viral genome sourced from recovered noninfectious patients would greatly facilitate epidemiological tracking. Thus, we developed a protocol to isolate and sequence the genome of SARS-CoV-2 from stool samples of post-acute SARS-CoV-2 patients, at time points ranging from 10-120 days after onset of symptoms.
Methods: Stool samples were collected from patients at varying time points post-convalescence and viral DNA was isolated and sequenced using the QIAamp viral RNA mini kit (Qiagen Inc.) and ion Ampliseq™ library kit plus (Life Technologies Corporation). Capacity of neutralizing antibodies in patient plasma was tested using a Luminex panel (Coronavirus Ig Total Human 11-Plex ProcartaPlex™ Panel, Thermo Fisher).
Results: Out of 64 samples obtained from post-acute patients, 21 (32.8%) yielded sufficient material for whole genome sequencing. This allowed us to identify widely divergent phylogenetic relativity of the SARS-CoV-2 genome from postacute patients living in the same households and infected around the same time. Additionally, we observed that individuals who recovered from infection expressed varying degrees of antibodies against SARS-CoV-2 structural proteins that corresponded to distinct variants. Interestingly, we identified a novel point mutation in the viral genome where infected patients expressed antibodies with a significantly reduced capacity to neutralize the virus in vitro relative to that of those infected with the wild type strain.
Conclusion: Altogether, we demonstrate a protocol to successfully sequence the SARS-CoV-2 genome from stool samples from patients up to 4 months post-infection, which can be applied to studies that assess the relationship between variants and immune response post-hoc and safe monitoring of the SARS-CoV-2 genome over the course of the pandemic.
Keywords: SARS-CoV-2; COVID-19; Variants; Sequencing; Stool; Diagnostic techniques; Mutations
Introduction
At the end of 2019, an outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), in Wuhan, China, quickly grew into the worldwide COVID-19 pandemic [1]. As of 10 May 2022, there have been over 500 million confirmed cases of COVID-19 globally, with over 6.25 million confirmed deaths, including one million in the U.S [2]. SARS-CoV-2 is thought to be a novel recombinant virus of the Coronaviridae family and similar to SARS-CoV and Middle East Respiratory Syndrome Coronavirus (MERS-CoV), has a zoonotic origin, likely bats or pangolins [3]. SARS-CoV-2 is a β-coronavirus, containing a ~29 kb positive-sense RNA genome, whose pathogenesis of human infection primarily affects the respiratory tract and varies in severity from mild symptoms to severe respiratory failure, concurrent with cough, fever, myalgia, dyspnea, headache and Gastrointestinal (GI) symptoms such as nausea and diarrhea [4-6].
Similar to SARS-CoV, SARS-CoV-2 relies on Angiotensin Converting Enzyme 2 (ACE2) as a receptor to invade human host cells [7]. Tissue/cell type specific variability in ACE2 expression contributes to SARS-CoV-2 tissue tropism. For example, Wang, et al. showed that SARS-CoV-2 RNA is best detected in bronchoalveolar lavage fluid specimens, although viral RNA has also been found in sputum, nasal and pharyngeal swabs, feces, and urine [8,9]. Interestingly, Li, et al. showed that while lungs express moderate levels of ACE2, higher expression may occur in the small intestine, testis, kidneys, heart, thyroid and adipose tissue [10]. While viral RNA has been detected in post-mortem myocardial tissues, viral load did not correlate with degree of symptomatic cardiac involvement [11]. Similarly, male reproductive tissues are thought to be potential targets of SARS-CoV-2, due to high ACE2 expression in sertoli, leydig and germ cells [12,13].
In addition to clinical studies, in vitro human organoid studies showed that SARS-CoV-2 can infect blood vessels, kidney, liver and small and large intestines [14-16]. Wang, et al. further showed that the live virus could be detected in feces, supporting the GI tract as a target organ system for SARS-CoV-2, suggesting a fecal route of transmission. Analogously, Lin, et al. detected viral loads in the esophagus, stomach, duodenum and rectum via endoscopic sampling, while Xiao, et al. showed that SARS-CoV-2 can infect and enter GI cells [17,18]. This group also detected the virus in feces of patients, despite negative respiratory samples, suggesting that the virus can potentially persist in the GI tract longer than in the nasopharynx and the respiratory tract.
Given the rapid clearance of SARS-CoV-2 from the nasopharyngeal after the first few weeks of infection, the persistence of the virus in the GI tract and stool samples may provide an alternative means to study viral variants genetically identified post hoc in non-infectious, post-acute individuals. A recent study demonstrated the feasibility of viral detection in stool samples of patients for as many as 77 days after infection, though more normally stool samples have returned positive results up to 33 days after a negative nasopharyngeal test [19,20]. However, whether the full viral genome remains intact in stool sample and amenable to sequencing has not yet been demonstrated. Herein, we describe a protocol to isolate and sequence the genome of SARS-CoV- 2 from stool samples of post-acute patients and demonstrate an example of how this data was applied to understanding the interindividual variability in the immune response to infection during convalescence.
Materials and Methods
Sample collection
All human subject studies were approved by the institutional review board of the university of Hawaii under protocol number 2020-00411. All participants provided informed consent during enrollment. In addition, informed consent for children under 18 years old was obtained from a parent and/or legal guardian. All methods were performed in accordance with the relevant guidelines and regulations. Data was collected from Oahu residents who had tested positive for COVID-19 during recruitment, but less than 3 months before the first point of data collection. Patients could only enroll in the study if they were considered noninfectious and “cured” by their physician. Men and women, aged 5 to 78 years old at the time of recruitment, were invited for weekly follow-up to the research clinic for 6 weeks, where blood samples were collected at each visit. During the 2nd week, patients also provided a stool sample and were measured for height and weight (for BMI calculation). All participants and their respective survey responses, were anonymized with a unique numerical ID, to ensure privacy. Data from patients that missed more than two appointments were excluded from the analyses. Informed consent forms were provided to all patients and assent was given before participation in the study.
Viral RNA extraction from stool samples
To clarify stool samples, 0.5 mL stool aliquots, preserved in RNAlater™ stabilization solution (Thermo Fisher Scientific, Inc., Vilnius, Lithuania), were added to 2.5 mL of 0.89% NaCl solution and centrifuged for 20 min at 4000 g. The supernatant was collected and filtered through an 0.2 μm syringe filter. Then, 2 mL of the filtrate was concentrated in Amicon® Ultra 2 mL centrifugal filters at 4000 g, in 10 min intervals, until ~140 μL remained in the column, which was then recovered by inverting the column and spinning at 1000 g for 2 min. The recovered sample concentrates were processed via the QIAamp viral RNA mini kit (Qiagen Inc., Valencia, CA, USA) for RNA extraction. Next, RNA (2 μL) was converted to cDNA using the SuperScript™ VILO™ cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA), following the product's directions. The final cDNA product was then diluted in RNase-free water (1:2).
Library preparation and sequencing
The ion Ampliseq™ library kit plus (Invitrogen) was used to amplify viral RNA extracted from stool samples. Following the amplification procedures indicated in the manufacturer’s protocol, PCR was performed in two pools, each including specific primer sets. After PCR amplification, full volume reaction products were run on e-gel precast agarose gels (Invitrogen). Successfully amplified PCR products were cut from the gel and pools 1 and 2 of each sample were combined into a single 1.5 mL tube. Then, PCR products were purified using GeneJET gel extraction kits (Thermo Fisher Scientific), following the manufacturer’s directions. To ensure sufficient amplification, a second PCR reaction was performed with the two primer pools using platinum™ PCR supermix high fidelity (Invitrogen). After the second amplification reaction, DNA libraries were prepared according to the ion Ampliseq™ library kit plus (Invitrogen) protocol. First, the amplicons were partially digested and adapters (Ion Xpress Barcodes Adapters 1-80 Kit, Invitrogen) were then ligated according to manufacturer’s protocol. Lastly, the libraries were purified using Agencourt™ Ampure™ XP beads (Beckman Coulter, Brea, CA, USA) and their concentrations were determined by qPCR using the Ion universal library quantification kit (Invitrogen), per the kit's instructions. Then, each library was diluted to a concentration of 39 pM and equal volumes of each library were pooled. The ion 510™ and ion 520™ and ion 530™ Kit (Thermo Fisher Scientific, Austin, TX, USA) was used to clonally amplify the pooled library on nanosized ionosphere particles by emulsion PCR. Bead enrichment and chip loading were also conducted using the Ion Chef Instrument. The parallel sequencing reaction was achieved on the ion S5 next generation sequencing system with ion 530 chips.
Luminex panel of anti-SARS-CoV-2 antibodies
Plasma collected from participants was evaluated against WT SARS-CoV-2 structural proteins using a Luminex panel (Coronavirus Ig Total Human 11-Plex ProcartaPlex™ Panel, ThermoFisher, Vienna, Austria), following the manufacturer’s protocol. The panel was read using a Luminex 200 instrument system (Thermo Fisher Scientific).
SARS-CoV-2 surrogate virus neutralization assay
SARS-CoV-2 surrogate Virus Neutralization Test (sVNT) kit (GenScript, NJ, USA) is a blocking ELISA which mimics the virus neutralization process, detecting circulating neutralizing SARS-CoV-2 antibodies that block the interaction between RBD and ACE2 on the cell surface receptor of the host. The test is isotype and species independent. Plasma samples were diluted 10X with sample dilution buffer and assayed following GenScript protocol. The absorbance of the sample is inversely dependent on the titer of the anti-SARS-CoV-2 neutralizing antibodies. S-RBD (wild type) was used in this assay.
Statistical analysis
Comparative analyses of immunological data in between different lineages were performed using One-way ANOVA test, followed by Tukey multiple comparison test. Unpaired t-test was used to evaluate impact of punctual mutations on inhibition assay. Graphing and statistical analysis were performed using Prism 9, Version 9.0°c (GraphPad, La Jolla, CA, USA). All statistical significance was determined at P<0.05.
Results
Recruitment, sample processing and sequencing: We recruited study participants during the first wave of SARS-CoV-2 infections in Honolulu, Hawaii, between June and September 2020. The individuals from whom samples were collected represented a range of ages, ethnicities and genders, with enrichment of the Native Hawaiians and other Pacific Islanders (NHPI) that comprises approximately 25% of the state’s population (Table 1).
| N | Number 67* | Percentage |
|---|---|---|
| Gender | ||
| Female | 40 | 60% |
| Male | 27 | 40% |
| Age (years) | ||
| Less than 30 | 19 | 28% |
| From 30 to 55 | 35 | 52% |
| Greater than 55 | 13 | 20% |
| Ethnicity | ||
| White | 21 | 31% |
| NHPI | 27 | 44% |
| Asian | 14 | 21% |
| Other | 5 | 4% |
| Major symptoms | ||
| Fever | 23 | 34% |
| Cough | 23 | 34% |
| Muscle Pain | 4 | 6% |
| Headache | 18 | 27% |
| Sore throat | 30 | 45% |
| Loss of taste or smell | 16 | 24% |
| Trouble breathing | 27 | 44% |
| HbA1C (% in blood) | ||
| Healthy (Bellow 5.7%) | 54 | 81% |
| Prediabetic (Between 5.7% and 6.4%) | 0 | 0% |
| Diabetic (Over 6.5%) | 8 | 12% |
| Didn’t want to get tested | 5 | 7% |
| Blood pressure category | ||
| Normal (systolic bellow 120, diastolic below 80) | 22 | 33% |
| Prehypertension (systolic between 120-139, diastolic between 80-89) | 31 | 46% |
| Hypertension (systolic above 140, diastolic above 90) | 7 | 10% |
| Didn’t want to get tested | 7 | 10% |
Table 1: Sociodemographic characteristics of total study participant population.
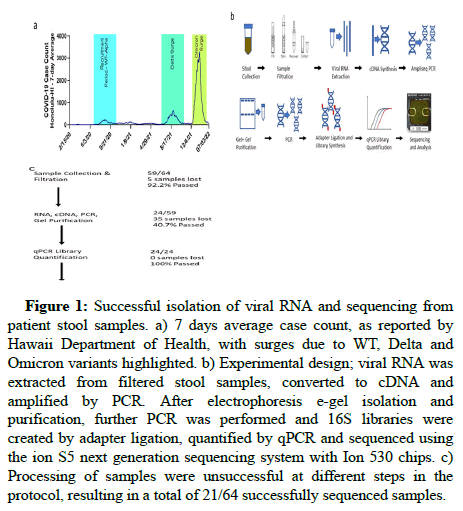
Figure 1a illustrates the 7 days average of reported cases over time in Hawaii, showing multiple surges in COVID-19 cases as a result of dominant variants, including delta and omicron, in relation to the alpha/WT variant surge suspected to have been prevalent during our recruitment period. Figure 1b illustrates the workflow used to amplify the SARS-CoV-2 RNA from stool samples. Of 64 donor stool samples, 21 (32.8%) were successfully sequenced, showing persistence of the viral genome in stool up to four months after infection (Figure 1c). Of the 43 samples that could not be sequenced, 5 failed the sample collection and filtration step, 35 failed to amplify after the first PCR step and 3 were lost following sequencing due to insufficient numbers of reads. All 21 sequenced samples exceeded quality control thresholds (Table 2). Sequenced libraries generated from these samples had an average of 91% mapped reads, 35X mean depth and a mean 85.6% quality score of Q20, indicating 99% base call accuracy. These data indicate high quality sequencing results.
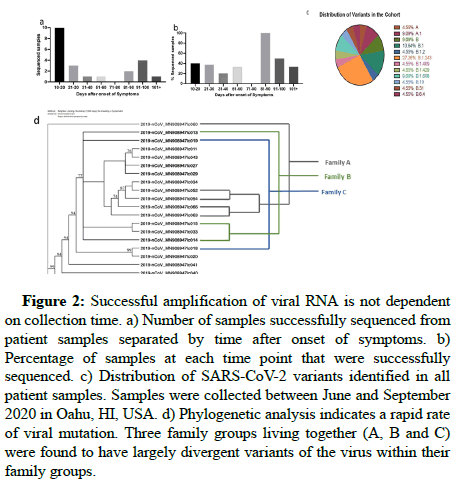
As shown in Figure 2, the largest absolute number of successfully sequenced SARS-CoV-2 genomes were from samples collected within a month after the onset of symptoms. However, the proportion of successfully sequenced genomes did not significantly depend on convalescence, as the highest percentage of successful sequencing was within the 81-90 day interval, with 100% of the samples sequenced. This suggests that time after infection may not be crucial for successful sequencing of the SARS-CoV-2 genome from fecal samples. The ability to amplify the samples may also depend on technical factors, such as sample storage condition and duration or biological variables such as individual viral load and shedding.
| Sample | Bases | Reads | Mean read length | Mapped reads | Mean depth |
|---|---|---|---|---|---|
| c009 | 1,198,205,462 | 6,324,538 | 189 | 6,044,096 | 36,938 |
| c010 | 1,489,569,391 | 7,998,512 | 186 | 7,659,778 | 46,434 |
| c011 | 1,420,405,554 | 7,782,617 | 182 | 7,351,799 | 43,158 |
| c013 | 1,265,373,919 | 7,130,732 | 177 | 6,785,235 | 37,972 |
| c014 | 896,894,202 | 5,145,390 | 174 | 5,009,620 | 27,482 |
| c015 | 1,402,887,604 | 7,838,674 | 178 | 7,629,091 | 43,051 |
| c020 | 1,094,646,127 | 6,366,095 | 171 | 6,134,237 | 33,530 |
| c019 | 1,258,534,760 | 7,055,747 | 178 | 6,932,524 | 40,648 |
| c018 | 1,268,627,314 | 6,923,648 | 183 | 6,642,191 | 37,989 |
| c029 | 1,250,649,554 | 7,022,447 | 178 | 6,719,011 | 37,883 |
| c027 | 1,833,125,595 | 10,128,995 | 180 | 9,407,724 | 52,309 |
| c060 | 521,208,581 | 2,975,609 | 175 | 1,895,676 | 2,749 |
| c063 | 1,045,161,100 | 6,184,851 | 168 | 4,970,854 | 29,312 |
| c066 | 999,836,546 | 5,145,831 | 194 | 5,042,641 | 31,993 |
| c061 | 817,345,387 | 4,406,087 | 185 | 2,536,814 | 105.4 |
| c033 | 1,360,821,401 | 7,702,775 | 176 | 6,762,678 | 32,315 |
| c034 | 905,566,710 | 4,983,346 | 181 | 3,296,075 | 20,372 |
| c040 | 979,524,142 | 5,468,628 | 179 | 5,293,151 | 30,302 |
| c041 | 1,457,526,880 | 7,898,404 | 184 | 7,654,353 | 46,282 |
| c043 | 1,176,684,729 | 6,547,919 | 179 | 6,463,230 | 38,207 |
| c052 | 1,529,825,053 | 7,889,273 | 193 | 7,848,024 | 50,528 |
| c054 | 1,670,225,938 | 8,761,159 | 190 | 8,694,724 | 54,565 |
| average | 1,220,120,270 | 6,712,785 | 181 | 6,216,978 | 35,187 |
| SEM | 64342038.53 | 340133.86 | 1.43 | 400391.9 | 2937.02 |
Table 2: Quality control of next generation sequencing.
Figure 1: Successful isolation of viral RNA and sequencing from patient stool samples. a) 7 days average case count, as reported by Hawaii Department of Health, with surges due to WT, Delta and Omicron variants highlighted. b) Experimental design; viral RNA was extracted from filtered stool samples, converted to cDNA and amplified by PCR. After electrophoresis e-gel isolation and purification, further PCR was performed and 16S libraries were created by adapter ligation, quantified by qPCR and sequenced using the ion S5 next generation sequencing system with Ion 530 chips. c) Processing of samples were unsuccessful at different steps in the protocol, resulting in a total of 21/64 successfully sequenced samples.
Figure 2: Successful amplification of viral RNA is not dependent on collection time. a) Number of samples successfully sequenced from patient samples separated by time after onset of symptoms. b) Percentage of samples at each time point that were successfully sequenced. c) Distribution of SARS-CoV-2 variants identified in all patient samples. Samples were collected between June and September 2020 in Oahu, HI, USA. d) Phylogenetic analysis indicates a rapid rate of viral mutation. Three family groups living together (A, B and C) were found to have largely divergent variants of the virus within their family groups.
Sequencing identified several SARS-CoV-2 subvariants present in the tested population, whose distribution is shown in Figure 2c, showing predominance of B1.243 (27.26%) and B.1 (13.64%) subvariants, earlier than reported by the state of Hawaii in May 2021 [21]. Our data was collected between June and September 2020, while the report describes data collected in the two weeks prior to the date shown, beginning in May 2021. According to the state report, 90% of all sequenced genomes at the time were subvariants B.1.429 and B. 1.1.7. During our recruitment period, B.1.429 only made up 4.55% of the subvariants present, while B.1.1.7 was fully absent from the population tested. However, in the few short months between September 2020 and May 2021, the dominant variants in the population changed and new variants emerged or predominated.
SARS-CoV-2 strains in families. To determine the relationship of viral subvariants within families, we performed phylogenetic analyses that revealed the viral genome to differ significantly within identified patient family clusters. Three patient families were identified, termed families A, B or C. Since individuals within each family lived together and reported concurrent symptoms, they were likely infected with SARS-CoV-2 at the same time, with the same or similar subvariant. However, genome data demonstrated sequentially divergent subvariants within each family cluster. For example, one family A member had a subvariant with 619 nucleotide changes relative to a subvariant found in another family A member. This may imply a rapid mutation rate or multiple concurrent subvariants in the population.
Plasma antibody neutralization of distinct viral strains. Based on prior studies showing interindividual differences in immune responses against SARS-CoV-2 [22] we explored how immune response may vary based on viral genotype. We first measured antibody levels against SARS-CoV-2 from plasma of post-acute patients in our study. The levels of antibodies against the nucleocapsid, spike, spike subunit 1 and RBD structural proteins varied across individuals in association with specific SARS-CoV-2 strains (Figure 3a-d). In contrast to the highest levels of antibodies observed against all structural proteins in patients infected with subvariants B1.608 and B.31, low to no antibodies were detected against these proteins in individuals infected with the B.10 and B.1.409 subvariants. Further, individuals recovering from infection with the B.1.409 and B.10 subvariants exhibited a lower in vitro neutralization capacity of SARS-CoV-2 than that of others (Figure 3e). In addition to these subvariants, we identified two novel missense mutations in the gene encoding the Nucleocapsid (N) protein of SARS-CoV-2. The mutation 419A>G had no effect on the capacity of antibodies from post-acute SARS-CoV-2 infected patients to neutralize the virus in vitro (data not shown). However, the mutation 1166A>G exhibited a reduced capacity to neutralize the virus in vitro by 30% relative to post-acute COVID-19 patients without this mutation (Figure 3f).
Figure 3: A novel point mutation in our cohort reduces neutralization ability of SARS-CoV-2 in patient plasma samples. a-d) Neutralization ability was assessed using patient plasma samples against SARS-CoV-2 nucleocapsid. a) Spike b) Spike S1 subunit. c) And receptor-binding. d) Domains, for each identified variant. Higher levels of neutralization indicated higher seroprevalence of SARSCoV-2 protein specific antibodies. e) Total inhibition ability against the entire virus was assessed by variant. Data shown is the average percentage of successful neutralization of each variant by patient plasma samples. f) The ability of patient plasma samples to neutralize a newly identified variant with a point mutation c.419A>G was compared to the WT variant. No significant difference was found in neutralization capacity due to this mutation (one-tailed unpaired t-test, p=0.4779). g) The ability of patient plasma samples to neutralize a newly identified variant with a point mutation 1166A>G was compared to the WT variant. A significant decrease in neutralization capacity was identified in the mutant, compared to common variant (one-tailed unpaired t-test, p=0.0115).
Discussion
The results of this study demonstrate the success and utility of a protocol to isolate and sequence the genome of SARS-CoV-2 from stool samples of post-acute patients, which can enable prospective studies to better understand the heterogeneity in the sequalae of postacute COVID-19. Although viral sequencing from fecal samples is a well-established technique in virology and has been used to characterize infections of many different viruses, our current study represents the first time a SARS-CoV-2 viral genome has been completely sequenced from stool samples of fully recovered patients. While previous studies have used other methods to sequence fecal samples from acute SARS-CoV-2 patients, our study uses a safer, easier and cheaper method than previously described [23–29]. Additionally, we demonstrate the possibility of high quality sequencing of the full SARS-CoV-2 genome for up to 120 days after infection. This indicates that samples may be collected without the use of a nasopharyngeal swab or the presence of active sickness or infection.
Although previous studies have demonstrated the utility of fecal swabs for early diagnosis or tracking viral circulation through wastewater, we demonstrate a utility for individual samples for evaluation in a prospective cohort of recovering patients [30,31]. This allows for better, less invasive tracking and characterization of various viral strains that emerge over the course of the pandemic [32-34]. Information collected in this manner may be used to improve strategies for COVID-19 mitigation strategies such as vaccination and quarantine. Indeed, studies reveal that more breakthrough cases may not only relate to waning vaccine induced immunity, but also associate with specific viral variants, indicating a clear need to continue monitoring changes in the distribution of variants over time and assess whether boosters, vaccines or anti-viral drugs may be warranted [35-39].
Conclusion
As an exploratory application of our method, we further examined the novel mutations we identified and the manner in which they might affect the activity of the virus. Prior studies show substantial interindividual variation in immune response against SARS-CoV-2. Although individual innate features such as genetics, gut microbiome, age, gender and obesity may contribute to this variability, it is unclear whether distinct mutations or viral subvariants also account for varying individual immune responses. We observed that individuals infected with distinct subvariants exhibited varying degrees of anti-SARS-CoV-2 antibody production. In particular, we found that SARS-CoV-2-recovering individuals carrying a novel mutation (1166A>G) in the Nucleocapsid (N) protein had significantly reduced viral neutralization capacity in vitro. We caution that these results are preliminary, as the cohort size was small, with asynchronous recovery periods from infection, altogether limiting our assessment of covariates (e.g., age) that may also contribute to interindividual differences in the immune response. However, these results support that genetic variation, intrinsic to SARS-CoV-2, may at least in part explain the interindividual variability in immune responses observed, warranting further investigation. These finding demonstrate the utility and relevance of our method in the context of a rapidly changing pandemic.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, AKM, upon reasonable request.
Competing Interests
The authors declare no competing interests.
Author Contributions
AKM and RJ obtained funding for the study. AKM, RP, RJ conceived of and designed the study. NP, NR, BK, LU, RL and NPA performed the sample collection, processing and neutralization assays. RP, NP, NPA wrote the manuscript and RP prepared the figures. All authors reviewed and approved the manuscript.
Funding
This study was supported in part by funding provided by Hawaii community foundation under award 20 HCF-101573. The comments expressed in this report are the sole responsibility of the authors and do not represent the official view of the Hawaii community foundation.
References
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China. N Engl J Med 382:727-733.
[Crossref] [Google Scholar] [PubMed]
- Johns Hopkins Coronavirus Resource Center (2022) COVID-19 Map. Johns Hopkins University and Medicine.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res 24:91-98.
[Crossref] [Google Scholar] [PubMed]
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270-273.
[Crossref] [Google Scholar] [PubMed]
- Hu B, Guo H, Zhou P, Shi ZL (2021) Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141-154.
[Crossref] [Google Scholar] [PubMed]
- Burke RM, Killerby ME, Newton S, Ashworth CE, Berns AL, et al. (2020) Symptom profiles of a convenience sample of patients with COVID-19 - United States, January-April 2020. MMWR Morb Mortal Wkly Rep 69:904-908.
[Crossref] [Google Scholar] [PubMed]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, et al. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454.
[Crossref] [Google Scholar] [PubMed]
- Wang W, Xu Y, Gao R, Lu R, Han K, et al. (2020) Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323:1843-1844.
[Crossref] [Google Scholar] [PubMed]
- Peng L, Liu J, Xu W, Luo Q, Chen D, et al. (2020) SARS-CoV-2 can be detected in urine, blood, anal swabs and oropharyngeal swabs specimens. J Med Virol 92:1676-1680.
[Crossref] [Google Scholar] [PubMed]
- Li M-Y, Li L, Zhang Y, Wang XS (2020) Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9:45.
[Crossref] [Google Scholar] [PubMed]
- Lindner D, Fitzek A, Brauninger H, Aleshcheva G, Edler C, et al. (2020) Association of cardiac infection with SARS-CoV-2 in confirmed COVID-19 autopsy cases. JAMA Cardiol 5:1281-1285.
[Crossref] [Google Scholar] [PubMed]
- Shen Q, Xiao X, Aierken A, Yue W, Wu X, et al. (2020) The ACE2 expression in Sertoli cells and germ cells may cause male reproductive disorder after SARS-CoV-2 infection. J Cell Mol Med 24:9472-9477.
[Crossref] [Google Scholar] [PubMed]
- Wang Z, Xu X (2020) scRNA-seq profiling of human testes reveals the presence of the ace2 receptor, a target for sars-cov-2 infection in spermatogonia, leydig and sertoli cells. Cells 9:920.
[Crossref] [Google Scholar] [PubMed]
- Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, et al. (2020) Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181:905-913.
[Crossref] [Google Scholar] [PubMed]
- Zhao B, Ni C, Gao R, Wang Y, Yang L, et al. (2020) Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell 11:771-775.
[Crossref] [Google Scholar] [PubMed]
- Lamers MM, Beumer J, van der Vaart J, Knoops K (2020) SARS-CoV-2 productively infects human gut enterocytes. Science 369:50-54.
[Crossref] [Google Scholar] [PubMed]
- Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, et al. (2020) Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 69:997-1001.
[Crossref] [Google Scholar] [PubMed]
- Xiao F, Tang M, Zheng X, Liu Y, Li X, et al. (2020) Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158:1831-1833.
[Crossref] [Google Scholar] [PubMed]
- Xiao F, Wan P, Wei Q, Wei G, Yu Y (2022) Prolonged fecal shedding of SARS-CoV-2 in a young immunocompetent COVID-19 patient: A case report and literature overview. J Med Virol 94:3133-3137.
[Crossref] [Google Scholar] [PubMed]
- Gupta S, Parker J, Smits S, Underwood J, Dolwani S (2020) Persistent viral shedding of SARS-CoV-2 in faeces-A rapid review. Colorectal Dis 22:611-620.
[Crossref] [Google Scholar] [PubMed]
- Hawaii Department of Health (2021) Hawaii sequencing and variants of SARS-Cov-2. Hawaii Department of Health.
- Pereira NL, Ahmad F, Byku M, Cummins NW, Morris AA, et al. (2021) COVID-19: Understanding Inter individual variability and implications for precision medicine. Mayo Clin Proc 96:446-463.
[Crossref] [Google Scholar] [PubMed]
- Deng L, Silins R, Castro-Mejía JL, Kot W, Jessen L, et al. (2019) A protocol for extraction of infective viromes suitable for metagenomics sequencing from low volume fecal samples. Viruses 11:667.
[Crossref] [Google Scholar] [PubMed]
- Bavelaar HHJ, Rahamat-Langendoen J, Niesters HGM, Zoll J, Melchers WJG (2015) Whole genome sequencing of fecal samples as a tool for the diagnosis and genetic characterization of norovirus. J Clin Virol 72:122-125.
[Crossref] [Google Scholar] [PubMed]
- Khalifeh A, Blumstein DT, Fontenele RS, Schmidlin K, Richet C, et al. (2021) Diverse cressdnaviruses and an anellovirus identified in the fecal samples of yellow-bellied marmots. Virology 554:89-96.
[Crossref] [Google Scholar] [PubMed]
- Strubbia S, Phan MVT, Schaeffer J, Koopmans M, Cotten M, et al. (2019) Characterization of norovirus and other human enteric viruses in sewage and stool samples through next-generation sequencing. Food Environ Virol 11:400-409.
[Crossref] [Google Scholar] [PubMed]
- Xu Y, Kang L, Shen Z, Li X, Wu W, et al. (2020) Dynamics of severe acute respiratory syndrome coronavirus 2 genome variants in the feces during convalescence. J Genet Genomics 47:610-617.
[Crossref] [Google Scholar] [PubMed]
- Cerrada-Romero C, Berastegui-Cabrera J, Camacho-Martínez P, Goikoetxea-Aguirre J, Pérez-Palacios P, et al. (2022) Excretion and viability of SARS-CoV-2 in feces and its association with the clinical outcome of COVID-19. Sci Rep 12:7397.
[Crossref] [Google Scholar] [PubMed]
- Kipkorir V, Cheruiyot I, Ngure B, Misiani M, Munguti J (2020) Prolonged SARS-CoV-2 RNA detection in anal/rectal swabs and stool specimens in COVID-19 patients after negative conversion in nasopharyngeal RT-PCR test. J Med Virol 92:2328-2331.
[Crossref] [Google Scholar] [PubMed]
- Mardian Y, Kosasih H, Karyana M, Neal A, Lau CY (2021) Review of current COVID-19 diagnostics and opportunities for further development. Front Med 8:615099.
[Crossref] [Google Scholar] [PubMed]
- Mohan SV, Hemalatha M, Kopperi H, Ranjith I, Kumar AK (2021) SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem Eng J 405:126893.
[Crossref] [Google Scholar] [PubMed]
- Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, et al. (2021) Vaccine breakthrough infections with SARS-CoV-2 variants. N Engl J Med 384:2212-2218.
[Crossref] [Google Scholar] [PubMed]
- Aleem A, Akbar Samad AB, Slenker AK (2021) Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19) statpearls. StatPearls Publishing.
[Google Scholar] [PubMed]
- Vasireddy D, Vanaparthy R, Mohan G, Malayala SV, Atluri P (2021) Review of COVID-19 variants and COVID-19 vaccine efficacy: What the clinician should know? J Clin Med Res 13:317-325.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Jazwinski SM (2018) The gut microbiota and healthy aging: A mini-review. Gerontology 64:513-520.
[Crossref] [Google Scholar] [PubMed]
- Bakhshandeh B, Sorboni SG, Javanmard AR, Mottaghi SS, Mehrabi MR, et al. (2021) Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect Genet Evol 90:104773.
[Crossref] [Google Scholar] [PubMed]
- Fricke-Galindo I, Falfán-Valencia R (2021) Genetics Insight for COVID-19 Susceptibility and Severity: A review. Front Immunol 12:622176.
[Crossref] [Google Scholar] [PubMed]
- Gao Y-D, Ding M, Dong X, Zhang JJ, Kursat Azkur A, et al. (2021) Risk factors for severe and critically ill COVID-19 patients: A review. Allergy 76:428-455.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Chi J, Lv W, Wang Y (2021) Obesity and diabetes as high-risk factors for severe coronavirus disease 2019 (Covid-19). Diabetes Metab Res Rev 37:e3377.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi