Research Article, J Vet Sci Med Diagn Vol: 12 Issue: 4
The Detection and Molecular Characterization of Canine Parvovirus Type-2 in Dogs with Gastroenteritis Symptoms in Ankara Province, Turkey
Sepandar Gargari* and Taner Karaoglu
Department of Virology, Ankara University, Ankara Province, Turkey
- *Corresponding Author:
- Sepandar Gargari
Department of Virology,
Ankara University,
Ankara Province,
Turkey;
E-mail: gargari@ankara.edu.tr
Received date: 25 August, 2020, Manuscript No. JVSMD-23-17992;
Editor assigned date: 28 August, 2020, PreQC No. JVSMD-23-17992 (PQ);
Reviewed date: 11 September, 2020, QC No. JVSMD-23-17992;
Revised date: 06 June, 2023, Manuscript No. JVSMD-23-17992 (R);
Published date: 04 July, 2023, DOI: 10.35248/2325-9590.23.12.100058
Citation:Gargari S, Karaoglu T (2023) The Detection and Molecular Characterization of Canine Parvovirus Type-2 in Dogs with Gastroenteritis Symptoms in Ankara Province, Turkey. J Vet Sci Med Diagn 12:4.
Abstract
Parvoviruses can cause important and different viral diseases and infections in human and animals. Canine parvovirus infection is one of the infectious diseases with acute, fibrinous, necrotic or hemorrhagic enteritis which is common in dogs around the world. First canine parvovirus 2 reported in America in 1978. As a result of the analysis of CPV isolates with the monoclonal antibodies and enzymes, a strain (CPV2a) with its new antigenic properties appeared in America in 1979. In the following years, CPV2b was detected in 1984 in America and CPV2c in 2001 in Italy. In this study, identification, molecular characterization of the virus that is circulating in dogs at the same time in Turkey, determine the dominant type of virus and identify the location of virus in the phylogenetic tree and finally virus isolation was aimed. For this purpose, 100 samples were taken from dogs with gastroenteritis symptoms, among them, 52 of them were identified as PCR positive. Fifteen positive products were taken for next generation sequence and subjected to phylogenetic analysis. Five samples were observed as: CPV-2a, 9 samples as: CPV-2b and 1 sample as: CPV-2c. Seventeen CPVs were isolated in this study. In the phylogenetic viewpoint, TR-04, TR-06, TR-10, TR-11 and TR-15 constructs were identified as CPV-2a according to this analysis and were founded to be closely genetics related with FJ005257 (Italy), GU362934 (Italy), FJ005259 (Italy), KF385389 (Italy) and KF385390 (Italy). TR-01, TR-02, TR-03, TR-07, TR-08, TR-09, TR12, TR13 and TR-14 were included in the phylogenetic tree as CPV-2b and were founded to be closely genetics related with KF373569 (Italy), FJ222823 (Italy), FJ005264 (Italy), DQ025992 (France), FJ005260 (Germany) and KP682525 (Spain). TR-05 were located as CPV-2c and were found to be closely genetics related with DQ025942 (France), FJ005235 (Italy), DQ02956 (France) and FQ005246 (France) and in the order of GQ865518 (Greece), FJ005247 (Belgium), FJ005214 (Spain), FJ005237 FJ222821 (Italy) sequences. Fourteen of these parvovirus isolates can’t made cytopathic effect in CRFK cell line but 3 of them had cytopathic effect in CRFK cells line. Finally, with IF test, virus isolates were stabilized as: CPV type 2.
Keywords: CPV-2; Isolation, Phylogenetic analysis, Sequencing, Gastroenteritis symptoms
Introduction
Canine Parvovirus (CPV) is a member of the genus protoparvovirus (family Parvoviridae, subfamily Parvovirinae) and together with Feline Panleukopenia Virus (FPLV) is now included in a unique viral specie, carnivore protoparvovirus 1. CPV is a small, non-enveloped, single-stranded DNA virus. Genome consists of an approximately 5,232-nucleotide. The whole genome of parvovirus involves the two open reading frames encoding for two Non-Structural (NS1/NS2) and two structural (VP1/VP2) proteins was amplified and identified. ORF1 and ORF2, encoding for Non-Structural (NS1 and NS2) and structural (VP1 and VP2) proteins. VP2 is the major capsid protein (about 1755 nt), represents the major determinant of parvovirus host range. For this reason, most studies during the years have focused on the evolution of the VP2 gene. Amino acid changes in VP2 protein which is the main viral capsid protein, causes the antigenic properties of the virus/host distribution and tissue tropism. Carefully characterization and identification of CPV type 2, needs more time, methods and sensitive techniques to be recognized [1].
CPV is a viral infection characterized by acute, fibrinous, necrotic or hemorrhagic enteritis in dogs and carnivores. Parvoviral enteritis is the usual form of the disease, especially in newborn puppies (4-8 weeks) by myocardial form, which is characterized by nonsuppurative myocarditis and leukopenia which causes sudden deaths. Canine minute virus or Canine Parvovirus (CPV-1) was isolated first time in 1967 from the feces of healthy dogs [2]. This isolate is different from Canine Parvovirus (CPV-2) which is spreaded in the world nowadays. After that, this disease was identified in many regions of the world and Turkey too. CPV-2 antigenically is different from the CPV-1 which causes pandemic hemorrhagic enteritis on dogs in the world. For the first time canine parvovirus-2 was reported in America in 1978. After the emergence of CPV-2, this virus was spreaded in world very quickly and it has become one of the important infectious agents for pet and wild dogs population. By Analysis of CPV-2 with using monoclonal antibodies and different enzymes, the new strain of CPV which was carrying the new antigenic properties, was identified in 1979 as CPV-2a. After that CPV-2a has been found in America in 1980-1981. As a result of the research on this isolated virus in the subsequent years, another antigenic variant of virus, (CPV-2b) was appeared in 1984-1986 which was replaced with CPV-2a in most area of USA. In the following years, the new variant CPV-2c was first seen in Italy [3].
In this study, identification, molecular characterization of the virus that is circulating in dogs at the same time in Turkey, determine the dominant type of virus, identify the location of virus in the phylogenetic tree and finally virus isolation was aimed [4].
Materials and Methods
Sample collecting
A total of 100 of parvovirus infection suspected samples with clinically symptomatic gastroenteritis were collected from private veterinary of clinics in Ankara (regardless of vaccination status) during 2013. The all tests of these samples were done between 2013-2014. According to gastroenteritis symptoms in this animals, 5 stool, 35 rectal swap, 3 nasal swap and 57 blood sample were collected (animal experiments were approved by the local ethics committee). Age distributions of sampled animals are: (0-3 mount, 17 numbers), (3-6 mount, 25 numbers), (>6 mounts, 26 number) and (unknown age group 32 number) [5].
Preparation of samples for tests
These samples were homogenized in 1/5 PBS (phosphate-buffered saline, pH 7.2) and then clarified by centrifuging at 3000 × rpm for 30 min. The supernatants were collected to use for the subsequent diagnostic tests (PCRs tests and virus isolation) [6].
Template DNA preparation
For all blood sample centrifuge were done at 3000 × rpm for 10 min, after that leukocyte of this blood collected to viral DNA extraction. Other samples were homogenized in 1/5 PBS (phosphatebuffered saline, pH 7.2) and then clarified by centrifuging at 3000 × rpm for 30 min. The supernatants were collected to viral DNA extraction. For viral DNA extraction, high pure viral nucleic acid kit was used (by using kit manual method) (high pure viral nucleic acid kit, Roche, German) [7].
Primer pair and PCR amplification
PCR tests were done by using pair primers PF (5- ATGGCACCTCCGGCAAAGA-3) and PR (5- TTTCTAGGTGCTAGTTGAG-3) of the CPV-2 gene encoding capsid protein: VP1 and VP2. The product of PCR these primers was 2245 bp. PCR’s were done by using solis biodyne hot firepol DNA polymerase. For PCR test thermal cycler conditions: Initial denaturation in 95°C for 10 min, denaturation in 94°C for 60 second, annealing in 52°C for 60 second, extension in 72°C for 150 second and final extension in 70°C for 10 min were done. After PCR test by using electrophoresis method, product was runned in 1% agarose gel (Prona, EU) in TAE (Tris Acetat Ethylene diamine tetra acetic acid, 5X) for 30 min [8].
Purification of PCR products and sequence and phylogenetic analysis
For this purpose, 20 μl of PCR product and 2 μl of 6 × loading dye were mixed and run in 1% gel, with 100 bp-3000 bp DNA marker (100 bp DNA ladder solis biodyne). PCR products that include 2245 bp were separated by cutting gel with the sterile bistoury and transferred into 1.5 ml eppendorf tubes. Then this gel pieces were purified by using commercial purification kit (high pure PCR clean/ extraction kit, Roche, Germany) by using the manufacturer's recommended method [9]. The obtained purified DNAs were stoked at 20°C and then used in sequence analysis. Fifteen amplified PCR products samples were selected for sequencing. After gel extraction and cleaning, they were sequenced to next generation sequencing by using the SBS method (sequencing by synthesis) and Miseq (Illumina Inc kit). For analysis of the data obtained after reading, the whole genome sequence of parvovirus (M19296) were used as references with using program IGV 2.3. The Unipro UGENE version 1.15.0 program was used to convert the data from BAM file to the fast from considering this data, the targeted 2245 bp region for 15 different products was cut off to get VP-2 region with 1755 bp. Because we wanted to perform our phylogenetic analysis only in VP-2 capsid protein region [10].
The 15 nucleotide sequences obtained in this study were aligned with the bioedite sequence alignment editor program. Then based on the changes in nucleotide sequences, 15 amino acid translations were made by using the bioedit sequence alignment editor program. After that the amino acids were aligned by using the same program. Finally, the 15 CPV, VP2 region nucleotide sequences obtained were subjected to neighbor joining analysis in order to compare with the sequences of different CPV genotypes from the gene bank, which are mostly close relative nearest geographical origin. Evolutionary distances were calculated by using the Tamura-Nei method. Evolutionary analysis was done with MEGA 6 program [11].
Cell culture and direct immunofluorescence test
CRFK (Crendall feline kidney) cell culture was used for virus isolation. This cell culture supported by virology department of veterinary faculty of Ankara university. CRFK cell cultures continuity was used by, DMEM (Dulbecco's Modified Eagle's Medium), 10% FBS (Fetal Bovine Serum negative from BVD) and antibiotics (100 units/ml penicillin G potassium and 0.5 mg/ml streptomycin sulfate) [12]. The original sample of PCR positive samples which stored previously in -20°C, were subjected to virus isolation in cell culture. After 4 or 5 days of virus inoculation to cell culture (it’s depended to cell culture conditions), inoculated cell cultures were collected (regardless of CPE occurs or not in cell cultures) and 5 blind passages were done. In the isolation studies, PCR tests were applied to each passage fluid to see presence of the virus [13].
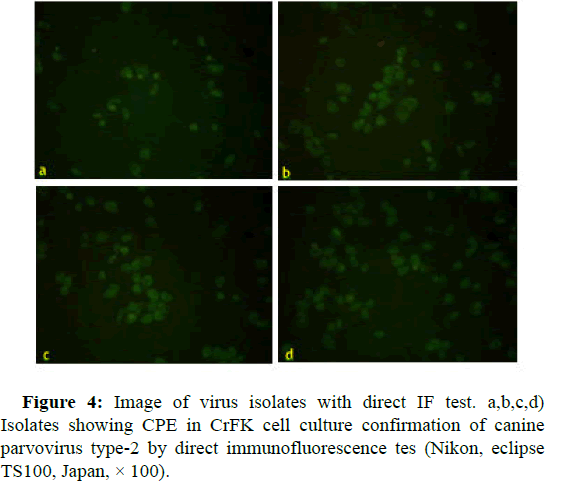
The primer wad used for molecular and phylogenetic analysis in this study, targets almost half of the virus genome. At the first stages of virus isolation studies, parvovirus titers which wanted to isolate, maybe was low. For following isolation studies, to perform positivity of presence of virus PCR were done. For this purpose, a primer that targets a small region of parvovirus partial capsid protein Vp2 was used. The product of these primers is 427 bp. In addition, the 5th passage fluids of serial passage the cell cultures and PCR positive of them, were subjected with Direct Immunofluorescence (DIF) test for the presence of canine parvovirus type 2. For immunofluorescence test, anti-CPV mouse monoclonal antibody labeled with Fluorescein Isothiocyanate (FITC) (VMRD, direct FA conjugate, CPV USA) was used. The conjugate only reacts only with CPV-2 [14].
Results
In the study, the presence of 2245 bp bands in the PCR result for stool, rectal swab, blood and nasal swab specimens for the VP-1 and VP-2 partial capsid protein gene regions were evaluated as positive for CPV was shows in Figure 1 [15].
A total of 52 dogs from 100 dogs were detected positive (52%) in terms of CPV. The distribution of PCR positivity according to the tested samples is presented in Table 1.
| Sample | Sample number | PCR (+) | PCR (+) % |
|---|---|---|---|
| Stool | 5 | 3 | 60 |
| Rectal swab | 35 | 20 | 57.1 |
| Blood | 57 | 28 | 49.1 |
| Nasal swab | 3 | 1 | 33.3 |
| Total | 100 | 52 | 52 |
Table 1: Distribution of PCR positive samples.
As a result of the PCR tests, 52 cases with positive CPV genotype DNA were analyzed by age. According to this, it was determined that 10 animals were 3 months old, 15 of the animals belong to 3-6 months old and 9 animals belong to more than 6 months old. Eighteen positive cases were unknown. Distribution of age group is presented in the Table 2.
| Age group | Sample number | PCR positive |
|---|---|---|
| 0-3 months | 17 | 10 |
| 3-6 months | 25 | 15 |
| 6> | 26 | 9 |
| Unknown | 32 | 18 |
Table 2: Age distribution of PCR positive sample.
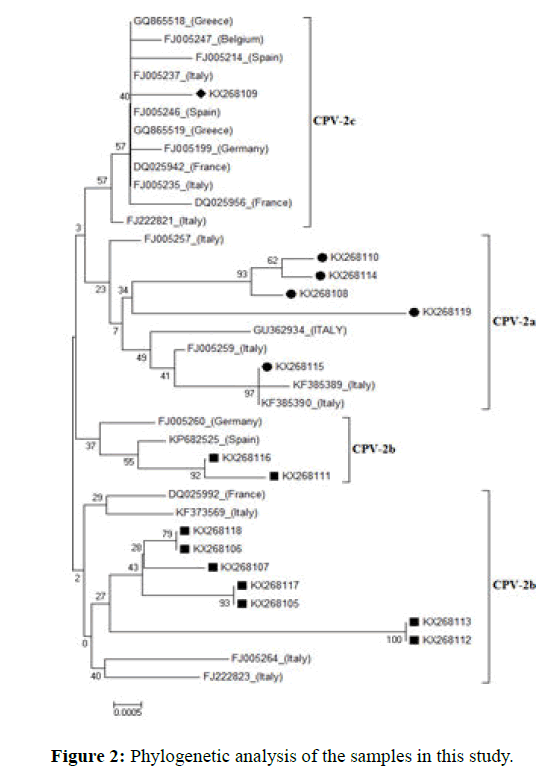
Fifteen were sequenced with next generation sequencing by using the SBS method (sequencing by synthesis) and Miseq (Illumina Inc kit). Neighbor-joining method use for phylogenetic analysis. The obtained phylogenetic analysis is presented in Figure 2 with phylogram format.
At the molecular level of the VP-2 region of CPV, 5 samples from 15 samples were observed CPV-2a, 9 samples were observed CPV-2b and 1 sample was observed as CPV-2c and took their place in the phylogenetic tree. These results were obtained that circulating virus in 2013, in Ankara mostly, CPV-2b (60%), CPV-2a (33.3%) and CPV-2c (6.6%) respectively (Figures 3 and 4).
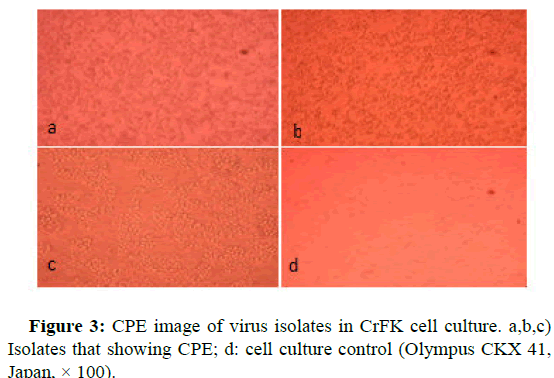
Twenty original samples with high PCR positivity from 52 PCR positive specimens were subjected to virus isolation in cell culture. By the way, all the samples transferred to the cell culture were prepared from 5 stool samples, 12 rectal swabs and 35 blood samples. After 5 blind passages in the cell culture, 17 samples were not produced CPE in the cell culture, while 3 samples were produced CPE.
The samples that cause CPE in the cell culture were subjected to a direct immunofluorescence test. The results of the direct IF test of 3 samples that showing CPE in cell culture were confirmed as canine parvovirus type 2.
Discussion
The examples in this study were collected in 2013. Amino acid change at position 297 (Ser-Ala) was observed both in CPV-2a and CPV-2b. Residue 297 is in a minor antigenic site close to epitope B and substitution at this position which might cause changes in CPV antigenicity variants. CPV-2a and 2b, have mutation in 297 position (Ser-Ala) which is designated as new CPV-2a and 2b. The aim of this study was molecular characterization and phylogenetic analysis of CPV and isolation canine parvovirus from fecal samples of dogs in CRFK cell line.
In the phylogenetic viewpoint, TR-04, TR-06, TR-10, TR-11 and TR-15 constructs were identified as CPV-2a according to this analysis and were founded to be closely genetics related with FJ005257 (Italy), GU362934 (Italy), FJ005259 (Italy), KF385389 (Italy) and KF385390 (Italy). In all CPV-2a in this study, mutations were occurred in the 426th position and asparagine amino acid, which was the result of the mutation at nucleotide in position 1276 (G→A). These types of dogs were found moderate/severe clinical table and prognosis in this type was good. It also has been reported scientifically that CPV-2a has a severe clinical table as CPV-2b, but the prognosis of the disease is good in most cases. Based on this, maybe the prognosis of the disease is good in this type. For this aim next research can be help. All of 5 CPV2-a detected, age groups, included in every (>6 months).
Similar findings have been reported previously in the US and China (12, 42). In one study in 2013, reported that CPV-2a is less than CPV-2b in Turkey. TR-01, TR-02, TR-03, TR-07, TR-08, TR-09, TR12, TR13 and TR-14 were included in the phylogenetic tree as CPV-2b and were founded to be closely genetics related with KF373569 (Italy), FJ222823 (Italy), FJ005264 (Italy), DQ025992 (France), FJ005260 (Germany) and KP682525 (Spain). In all these cases, similar CPV-2b in position 426, aspartic amino acid was found, except TR-08 and TR-09. Since the phylogenetic analysis carried out in this study at nucleotide level, mutation at 18 position (A→T) in TR-08 and TR-09 look like in KF373569 (Italy), FJ222823 (Italy), FJ005264 (Italy), DQ025992 (France), FJ005260 (Germany) and KP682525 (Spain) sequences, at the 147th position (G→A) the mutation look like FJ005260 (Germany), FJ222823 (Italy) and KP682525 (Spain) sequences and finally In the 426th position (A→G) mutation look like in DQ025992 (France) sequence were located as CPV-2b. Also, when we look at the clinical symptoms of dogs that are infected with this variety, moderate disease chart and less than of CPV-2a were shown. This situation has been reported scientifically that CPV-2a has a severe clinical table as CPV-2b. Odd thing, from 9 CPV2-b were identified, 4 dogs unresponsive and eventually died. This suggests that the disease has a severe clinical symptom (bloody diarrhea) especially in dogs that were infected with this variety. These 9 detected CPV2-b age groups were 6 dogs (3-6 months) and 3 dogs (>6 months).
The age of the sick dogs was mostly determined (3-6 months) which is consistent with the results of study in 2007 (30). TR-05 were located as CPV-2c and were found to be closely genetics related with DQ025942 (France), FJ005235 (Italy), DQ02956 (France) and FQ005246 (France) and in the order of GQ865518 (Greece), FJ005247 (Belgium), FJ005214 (Spain), FJ005237 FJ222821 (Italy) sequences. The other name for CPV-2c is 426-Glu. As a result of the mutation at position 1278 of TR-05 (T→A) and at the 426th a (asparagine amino acid→glutamic amino acid) was changed, which means that genetic proximity like other European CPV-2c as in TR-05. Furthermore, this detected CPV2-c belongs to an 8-year-old male kangal race dog. This dog recovered after several days of supplement treatment and the prognosis of the disease was good. When we look for this infected dog's disease table, the clinical symptoms of the disease were found moderate in this type. Although CPV infection is not often seen in young dogs, it has recently become a problem in adult dogs. There are several scientific reports on the presence of CPV-2c in adult dogs. Infection with CPV-2c was seen in a 12-yearold dog with fever (40.5°C), pulse and respiratory acceleration, vomiting and bloody diarrhea and lymphopenia within a few days, however, the dog was self-healing.
Conclusion
Parvoviruses can cause one of the important infectious diseases in dogs/ cats. Although parvoviruses are DNA viruses, has extremely high substitution rate. The mutation which see in this and seemlier study show this virus continuous can be change in structural proteins and causes to appear new viruses as well as RNA viruses. Because of study on this virus in next project, thought to be important in terms of disease and vaccine studies. This study is also the first isolation of CPV-2 in cell culture in Turkey. These viruses isolates can be help in vaccine and serologic study in future.
References
- Appel MJ, Scott FW, Carmichael LE (1979) Isolation and immunisation studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Veter Rec 105: 156-159.
[Crossref] [Google Scholar] [PubMed]
- Binn LN, Lazer BC, Eddy GA (1970) Recovery and characterization of a minute virus of canines. Infect Immun 1: 503-508.
[Crossref] [Google Scholar] [PubMed]
- Bounavoglia C, Martella V, Pratelli A (2001) Evidence for evolution of canine prvovirus type 2C in Italy. J Gen Virol 82: 3021-3025.
[Crossref] [Google Scholar] [PubMed]
- Carmichael LE (1994) Canine parvovirus type-2. An evolving pathogen of dogs. Ann Vet Med 135: 459-464.
- Carmichael LE (1999) Neonatal viral infections of pups: Canine herpesvirus and minute virus of canines (canine parvovirus 1). Rec Advanc Canine Infect Dis.
- Cavalli A, Bozzo G, Decaro N (2001) Characterization of a canine parvovirus strain isolated from an adult dog. New Microbiol 24: 239-242.
[Google Scholar] [PubMed]
- Cesar ADP, Telma AM, Dolores UM (2000) Molecular characterization of canine parvovirus in Brazil by polymerase chain reaction assay. Vet Microbiol 75: 127-133.
[Crossref] [Google Scholar] [PubMed]
- Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, et al. (2014) The family Parvoviridae. Archiv Virol 159: 1239-1247.
[Crossref] [Google Scholar] [PubMed]
- Csagola A, Varga S, Lorincz M, Tuboly T (2014) Analysis of the full-length VP2 protein of canine parvoviruses circulating in Hungary. Archiv Virol 159: 2441-2444.
[Crossref] [Google Scholar] [PubMed]
- Decaro N, Cirone F, Desario C (2009) Severe parvovirus in a 12-yearold dog that had been repeatedly vaccinated. Vet Rec 164: 593-595.
[Crossref] [Google Scholar] [PubMed]
- Decaro N, Desario C, Elia G (2008) Evidence for immunisation failure in vaccinated adult dogs infected with canine parvovirus type 2c. New Microbiol 31: 125-130.
[Google Scholar] [PubMed]
- Decaro N, Buonavoglia C (2012) Canine parvovirus a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol 155: 1-12.
- Dei GS, Cubeddu T, Giagu A, Sanna G, Rocca S, et al. (2017) First molecular characterization of canine parvovirus strains in Sardinia, Italy. Arch Virol 17: 3457-3463.
[Crossref] [Google Scholar] [PubMed]
- Hall TA (1999) BioEdit: A user-friendly biological sequence alignment and analysis program for windows 95/98/NT. Nucleic Acids Symp 41: 95-98.
- Hirayama K, Kano R, Hosokawa-Kanai T (2004) VP2 gene of a canine parvovirus isolate from stool of a puppy. J Vet Med Sci 67: 139-43.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi