Research Article, J Appl Bioinforma Comput Biol Vol: 7 Issue: 1
ADME/T Prediction, Molecular Docking, and Biological Screening of 1,2,4-Triazoles as Potential Antifungal Agents
Usman Rasheed1*, Umair Ilyas2, Shahiq Uz Zaman2, Syed Aun Muhammad3, Hasan Afzaal2 and Reem Altaf4
1Comsats Institute of Information and Technology, Abbot Abad, Pakistan
2Riphah Institute of Pharmaceutical Sciences, Riphah International University, Islamabad, Pakistan
3Department of Biotechnology, Bahaudin Zikriya University, Multan, Pakistan
4Shifa Tameer-e-Millat University, Islamabad, Pakistan
*Corresponding Author : Usman Rasheed
Comsats Institute of Information and Technology, Abbot Abad, Pakistan
Tel: +92 51 9247000
E-mail: samipharm@gmail.com
Received: August 22, 2017 Accepted: January 20, 2018 Published: January 27, 2018
Citation: Rasheed U, Ilyas U, Zaman SU, Muhammad SA, Afzaal H, et al. (2018) ADME/T Prediction, Molecular Docking, and Biological Screening of 1,2,4-Triazoles as Potential Antifungal Agents. J Appl Bioinforma Comput Biol 7:1. doi: 10.4172/2329-9533.1000144
Abstract
Clinical needs for new antifungal agents have gradually and steadily increased with the rise of AIDS-related mycoses, and the change in spectrum of fatal disseminated fungal infections.Triazoles are taken as potential antifungal molecules considering the existing portfolio of antifungal agents available in the market. This study was aimed to evaluate antifungal potential of 4-Amino 5-(2-Hydroxyphenyl)-1,2,4-Triazole-3-Thione and4-(2-hydroxybenzalidine)amine-5-(2-hydroxy)phenyl-1,2,4-triazole-3-thiol using molecular docking and in-vitro antifungal screening approach. This study was further designed to evaluate potential drug likeness properties and ADME/T prediction of aforementioned experimental triazoles. 3D crystal model of cytochrome P450 lanosterol 14 α-demethylaseenzyme was acquired from Protein Data Bank (PDB ID 5EQB). Docking studies revealed that both experimental compounds showed stable binding complex with lowest binding affinity values compared to reference standard Fluconazole (-6.8, -7.8 and -7.1Kcal/mol respectively for UI, UIA and Fluconazole) against cytochrome P450 lanosterol 14 α-demethylase, which is a key yeast target.In-vitro evaluation revealed that both experimental triazoles especially UIA showed promising MIC values, 24, 48 and 80 μg against Candida albican, Candida tropicalis and Candida glabrata respectively. Rat acute toxicity prediction using GUSAR model suggested that both experimental molecules were virtually non toxic when computationally studied through IV, IP, SC and oral routes of administration. This experimental work concludes that both triazoles possess strong fungicidal potential and should be further explored. In-silico work in current study further proposes an exceptional strategy for drug discovery with minimum cost and time
Keywords: 1,2,4 Triazoles; Antifungal; Computer aided drug designing; In-vitro; Molecular docking
Introduction
Invasive, life-threatening fungal infections are an important cause of morbidity and mortality, particularly for patients with compromised immune function. The number of therapeutic options for the treatment of invasive fungal infections is quite limited when compared with those available to treat bacterial infections. Indeed, only three classes of molecules are currently used in clinical practice and only one new class of antifungal drugs has been developed in the last 30 years [1] As a matter of fact, arsenal of antifungal drugs is relatively unfilled, with fewer numbers of fungicidal molecules commercially available compared to antibacterial agents. At parallel emergence of resistant fungal strains is alarmingly increasing day by day. Frightening boost in fungal infections is mainly linked to distressing increase in reporting of AIDS, cancer and organ transplant patients. Since treatment of aforementioned ailments lead to compromised immune status, so the patients are always exposed to opportunistic fungal infections. Especially amplified incidence of AIDS is another gigantic challenge in which patient is at risk of catching up fungal infections because of the well-known fact that patients having AIDS are with weakened immune status [2]. These clinical challenges have amplified the consumption of antifungal agents which ultimately is resulting in emergence of drug resistance within leading pathogenic fungal strains. Top culprit responsible for systemic and local candidiasis includes, Candida albican, followed by Candida glabrata and Candida tropicalis isolated from patient samples in clinical settings. Candida albican is frequently responsible for oral candidiasis (oral thrush), vaginal candidiasis, and candidemia [3,4]. Candida glabrata has become clinically significant because of increasing prevalence globally, as it is second leading organism responsible for candidemia [5]. Candida tropicalis has been identified as most prevalent Candida non-albican group of yeasts. This pathogen can be proclaimed as emergent pathogenic fungal specie as it’s reporting in causing Candidiasis has been dramatically increased [6]. Triazoles are biologically active moieties having various pharmacological properties including anti-inflammatory [7], analgesic [8,9] anti-cancer [10,11], anti-oxidant [12], anti-bacterial [13] and anti-fungal [14]. Many experimental 1,2,4 triazoles have shown promising antifungal potential in in-vitro and in-silico models [15-17]. Triazoles are backbone of currently available antifungal regimen. Most of commercially available antifungal molecules are triazoles in nature, including fluconazole, itraconazole and ketoconazole [18]. These triazoles target cytochrome P 450 dependent lanosterol 14 α-demethylase which is a key fungal target invovled in biosynthesis of sterols those are essential biochemical moeities responsible for carrying out various biological activitites within fungal cell. Its inhibition by different reported antifungal agents result in subsequent loss of normal sterols correlates with accumulation of 14 α-methyl sterols in fungi ultimately resulting in cell death [19].
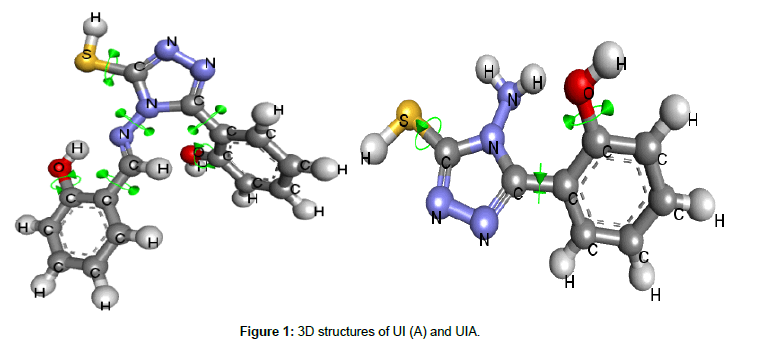
In current study we evaluated two triazoles i.e. 4-Amino 5-(2-Hydroxyphenyl)-1,2,4-Triazole-3-Thione and 4-(2-hydroxybenzalidine)amine-5-(2-hydroxy)phenyl-1,2,4-triazole- 3-thiol (Figure 1) for their antifungal potential. These experimental triazoles were already synthesized and characterized in Pharmaceutical Chemistry Lab of Riphah institute of pharmaceutical sciences [20], and were received as a gift for this experimental work. Evaluation of these triazoles was planned in three steps i.e. ADME/T prediction of experimental triazoles, docking studies against Cytochrome P 450 dependent lanosterol 14 α-demethylase enzyme and finally by In-vitro evaluation for antifungal activity.
Experimental Procedure
Prediction of physicochemical and ADMET properties
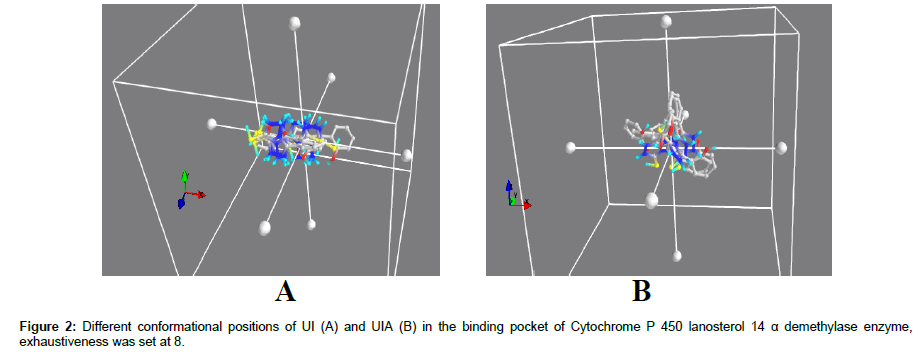
Phsyicochemical properties of both lead molceules were determined by using virtual predition tool FAF Drug 3 server [21]. UI and UIA compounds were screened through molinspiration online tool inorder to obtain their drug likeness characteristics. Rule of 5 also helps us to understand ADME/T properties of ligands. According to Rule of 5, ligand must carry following moecular characterisitcs inorder to be considered as a drug molecule: Hydrogen Bond Donor ≤5 (OH and NH Groups), Hydrogen Bond Acceptor ≤ 10 (N and O atoms), molecular weight ≤ 500 dalton and partition coefficient (Clogp) less than 5 [22,23] (Figure 2).
Toxicity prediction using GUSAR model: LD50 values for each lead molecules through different route of adminsteration including intravenous (IV), oral, subcutaneous (SC) and intraperitneal (IP) were predicted by using acute rat toxicity Generally Unrestricted Structure Activity Relationship (GUSAR) model [24].
Docking studies
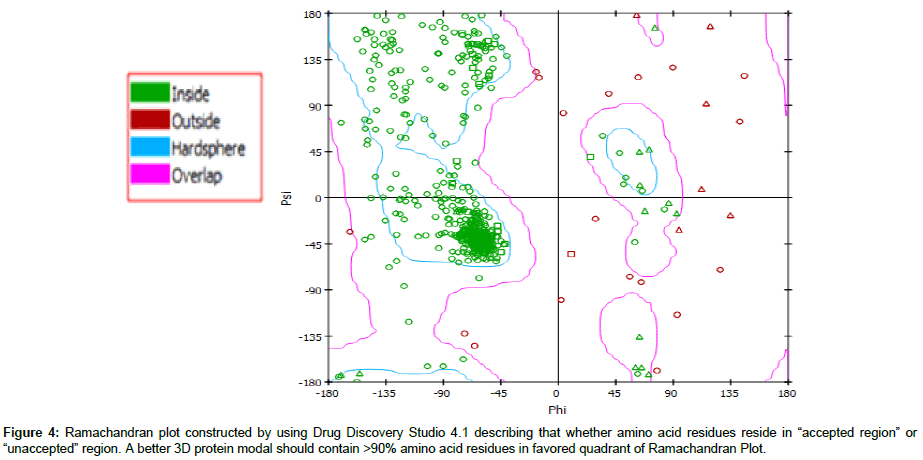
Molecular modeling: Structures of both triazoles UI and UIA were drawn and cleaned into 3D models using Marvin Sketch Tool and were saved in PDB formats [25] (Figure 1). Mol formats of both ligand molecules were converted into PDB files using Discovery Studio and were finally prepared into PDBQT file using PyRx tool inorder to produce atomic coordinates. 3D crystal model of cytochrome P450 lanosterol 14 α-demethylase enzyme (CaCYP51) was acquired from Protein Data Bank (PDB ID 5EQB) (Figure 2). Using Drug Discvoery Studio, Ramachandran Plots for the downloaded protein model. Ramachandran Plot tells about specific low energy conformations for Õ (phi) and (psi) or stable conformations of amino acid residues along with favorable and unfavorable regions for amino acid residues in plot. (Figure 3) Points on the plot indicate the Õ (phi) and (psi), the torsion angles of amino acid residues in a 3D protein model [26]. Both target protein (CaCYP51) and ligand were uploaded in BioLip Virtual Tool in order to find out the coordinates of ligand in original target protein site [27]. BioLip is an online Ligand-Protein binding database used for prediction of ligand binding residues in a target protein.
Docking run: Docking is a computational tool to evaluate interactiuon of target with ligand. To closely monitor and evaluate possible residual interaction between ligand and target protein, both ligand and macromolecule (target protein CaCYP51) were uploaded in PyRx Virtual Screening tool which is an Autodock Vina option based on scoring functions. Residual interaction of triazoles with target enzyme was evaluated by viewing different poses of interaction using Discvoery Studio [28].
Antifungal susceptibility testing
In-vitro antifungal suceptibility testing was done using serial dilution method inorder to find out minimum inhibitory concentration values (MIC) against C. albicans, C. glabratra and C. tropicalis [29].
Results
Prediction of physicochemical and ADMET properties
Predicted water solubility values (in mg/L) showed that UI showed comparable water solubility i.e., 31681.67 mg/L Vs. 38565.40 mg/L. On VEBER and EGAN models, both UI and Fluconazole were proposed to have good oral bioavailability profile. With solubility 8704.71 mg/L UIA was predicted as having less oral bioavailability compared to both UIA and Fluconazole (Table 1). Both triaozles UI and UIA fulfilled criteria of Rule of Five without any violation. Molecular weight for UI was 208 and 312 Dalton for UIA. Similary all other pharmacokinetic parameters (ADME/T) were between normal values for all lead molecules with zero violations which revealed both triazles can be developed as drug molecules (Table 1).
| Physicochemical Properties | UI | UIA | Fluconazole |
|---|---|---|---|
| Molecular Weight | 210.25 | 312.34 | 306.27 |
| Logp | 1.0 | 2.59 | 0.5 |
| LogD | 0.92 | 3.35 | 0.56 |
| logSw | 1.89 | 3.58 | -2.07 |
| Topological Polar Surface Area | 76.90 | 83.52 | 81.65 |
| Rotatable Bonds | 1 | 3 | 5 |
| Rigid Bonds | 11 | 18 | 16 |
| Flexibility | 0.083 | 0.14 | 0.24 |
| Hydrogen Bond Donor | 4 | 2 | 1 |
| Hydrogen Bond Acceptor | 5 | 6 | 7 |
| Rings | 2 | 3 | 3 |
| Max size ring | 6 | 6 | 6 |
| Num Charges | 0 | 1 | 0 |
| Total Charge | 0 | -1 | 0 |
| Heavy atoms | 14 | 22 | 22 |
| Carbons Atoms | 8 | 15 | 13 |
| Hetero Atoms | 6 | 7 | 9 |
| Ration H/C | 0.75 | 0.46 | 0.69 |
| Lipinski Violations | 0 | 0 | 0 |
| Solubility mg/L | 31681.67 | 8704.71 | 38565.40 |
| Solubility Forecast Index; | Good | Reduced Solubility | Good Solubility |
| Oral_Bioavailability_VEBER | Good | Acceptable | Good |
| Oral_Bioavailability_EGAN; | Good | Acceptable | Good |
Table 1: Physicochemical properties/ADME prediction.
Toxicity prediction using GUSAR model: Predicted LD50 values (in mg/kg) through IV, oral, SC and IP routes showed that both experimental triazoles are proposedly non toxic and thus can be further studied for their toxicity profile in animal models [30]. Toxicity prediction using GUSAR model further showed that LD50 values of UI and UIA were comparable with reference standard Fluconazole (Table 2)
| Route of Administration | LD50 mg/kg | ||
|---|---|---|---|
| UI | UIA | Fluconazole | |
| Intra-peritoneal | 280,900 | 481,900 | 708,400 |
| Intravenous | 64,320 | 193,600 | 200,400 |
| Oral | 764,600 | 1,548,000 | 584,000 |
| Subcutaneous | 521,990 | 1,212,000 | 511,300 |
Table 2: Rat acute toxicity values of selected ligand molecules using GUSAR.
Docking studies
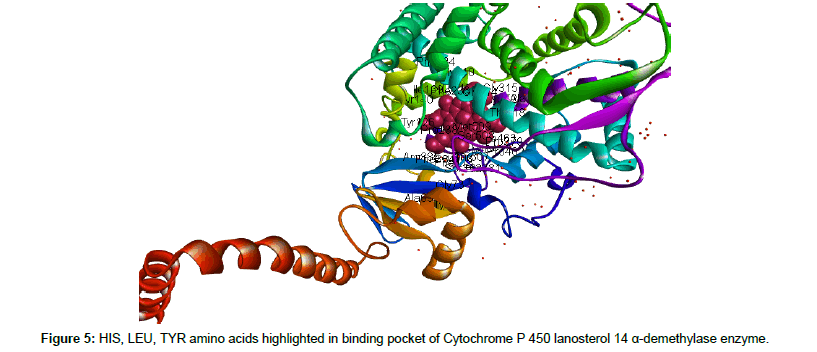
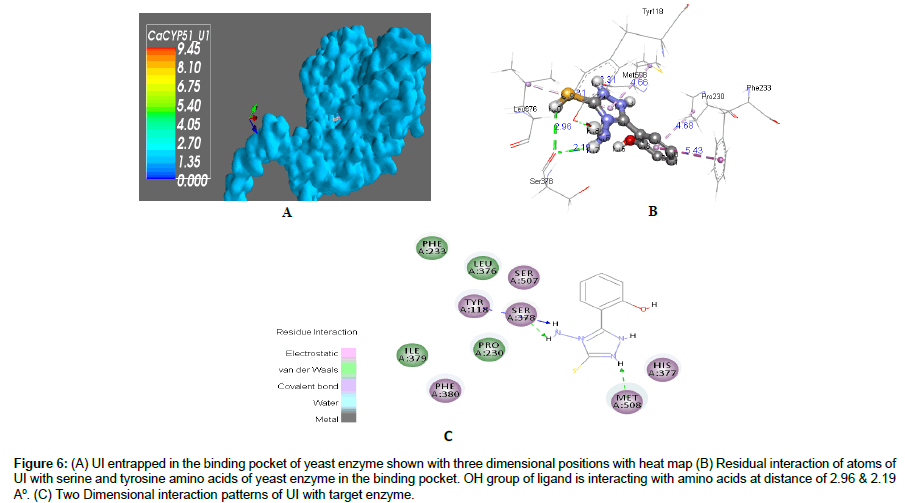
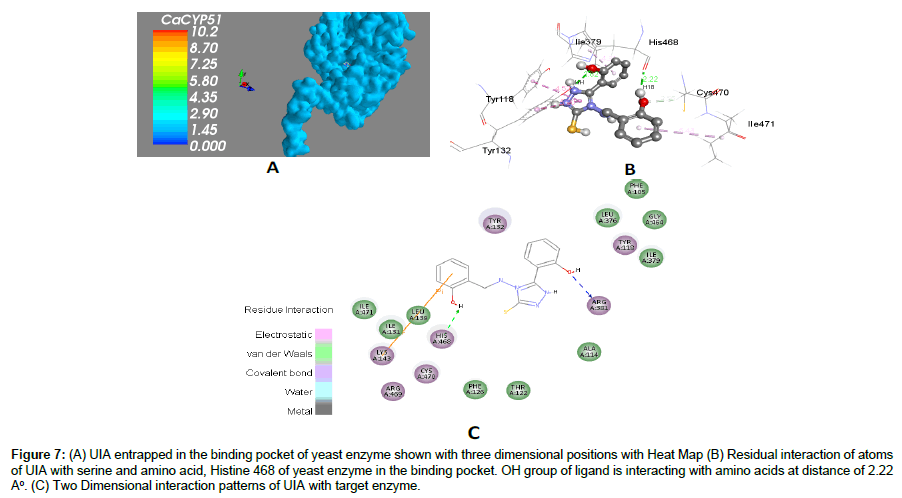
Binding site with atomic coordinates of ligand was analyzed by using online server 3DLigandSite. HIS, LEU and ALA amino acids were identified in the active binding pocket of target enzyme CaCYP51 (Figures 4 and 5). Docking interaction of both ligand against target protein (CaCYP51) revealed lowest binding affinity values indicating stable Ligand-Target complex which is a predictor of insilico inhibition of CaCYP51. Docking energies calculated using PyRx based on Autodock Vina were, -7.8 and -5.9 Kcal/mol with favorable RMSD values compared with -7.1 Kcal/mol with reference standard fluconazole (Table 3). Hydrogen interactions between ligand and target protein is an important predictor of strength of interaction between them which was also closely studied in our study and hydrogen bond donor and recepeint areas of both ligand and target was closely viewed as well as distance of interacting atoms were also monitored closesly (Figures 6 and 7).
Figure 6: (A) UI entrapped in the binding pocket of yeast enzyme shown with three dimensional positions with heat map (B) Residual interaction of atoms of UI with serine and tyrosine amino acids of yeast enzyme in the binding pocket. OH group of ligand is interacting with amino acids at distance of 2.96 & 2.19 A°. (C) Two Dimensional interaction patterns of UI with target enzyme.
Figure 7: (A) UIA entrapped in the binding pocket of yeast enzyme shown with three dimensional positions with Heat Map (B) Residual interaction of atoms of UIA with serine and amino acid, Histine 468 of yeast enzyme in the binding pocket. OH group of ligand is interacting with amino acids at distance of 2.22 A°. (C) Two Dimensional interaction patterns of UIA with target enzyme.
| Ligand-Target Complex | Binding Affinity | RMSD/UB | RMSD/LB | Auto dock Elements |
|---|---|---|---|---|
| Q5AEK7_CANAL_U1 | -6.8 | 0 | 0 | A, NA, OA, N, SA, HD |
| Q5AEK7_CANAL_U1 Q5AEK7_CANAL_U1 |
-6.1 -6.1 |
1.399 2.308 |
1.338 1.818 |
|
| Q5AEK7_CANAL_U1 | -5.6 | 2.026 | 1.371 | |
| Q5AEK7_CANAL_U1A | -7.8 | 0 | 0 1.338 |
A, C, OA, N, SA, HD |
| Q5AEK7_CANAL_U1A Q5AEK7_CANAL_U1A |
-7.0 -6.1 |
1.313 2.491 |
2.114 3.925 |
|
| Q5AEK7_CANAL_U1A | -6.1 | 3.383 | 0 | |
| Q5AEK7_CANAL_Fluconazole | -7.1 | 0 | 2.722 | A, C, F, OA, N, HD |
| Q5AEK7_CANAL_Fluconazole | -7.0 | 5.471 | 2.781 | |
| Q5AEK7_CANAL_Fluconazole | -6.9 | 5.625 | 3.290 | |
| Q5AEK7_CANAL_Fluconazole | -6.9 | 5.225 |
Table 3: Binding affinity energies and root Mean Square Deviation (RMSD) values obtained after docking run of ligands with target protein.
Antifungal susceptibility testing
In-vitro antifungal susceptibility testing that all fungal strains including C. albican, C. tropicalis and C. glabrata were susceptible to both experimental triazoles and with serial dilution method UIA showed better MIC values than UI (Table 4). UIA showed least MIC values against C. albicans i.e., 24μg which unveils its promising potential as antifungal agent and requires further investigation in in-vivo models.
| Organisms | MIC Values (μg) | ||
|---|---|---|---|
| UI | UIA | Fluconazole | |
| Candida albicans | 42 | 24 | 5 |
| Candida tropicalis | 44 | 48 | 10 |
| Candida glabrata | 98 | 80 | 8 |
Table 4: Minimum inhibitory concentration values.
Conclusion
Both lead molecules (UI and UIA) complied with Lipinski Rule of Five with zero violation reflecting that both ligand molecules are excellent drug candidates. Docking studies suggested that both lead triazoles including UI and UIA showed excellent binding affinity values especially UIA showed most stable ligand-target complex hence we propose UIA as an excellent inhibitor of Cytochrome P 450 dependent lanosterol 14 α-demethylase with compareable docking energy values with reference standard fluconazole. Rat acute toxicity prediction using GUSAR model suggested that both experimental molecules were nontoxic when computationally studied through IV, IP, SC and oral routes of administration. Docking outcomes were validated by performing in-vitro antifungal susceptibility testing of both UI and UIA. Antifungal susceptibility testing revealed that all three pathogenic fungal strains were susceptible to both triazoles. UIA showed even promising MIC values compared with fluconazole against Candida albican. Furthermore, Insilico studies propose an outstanding framework to screen small molecules for their possible biological activities in dry lab in minimum cost and time.
References
- Roemer T, Krysan DJ (2014) Antifungal drug development: challenges, unmet clinical needs, and new approaches.Cold Spring Harb Perspect Med 4: a019703.
- Vanden BH, Warnock DW, Dupont B, Kerridge D, Sen Gupta S, et al. (1994) Mechanisms and clinical impact of antifungal drug resistance. J Med Vet Mycol 32: 189-202.
- Richardson MD (2005) Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother 56: 5-11.
- Molero G, Díez-Orejas R, Navarro-García F, Monteoliva L, Pla J, et al. (2010) Candida albicans: Genetics, dimorphism and pathogenicity. Int Microbiol 1: 95-106.
- Tawanda G, Carlos IM, Géraldine H, Matthew KT, Steven MG (1999) Candida glabrata fungemia Clinical features of 139 patients. Medicine 78: 220-227.
- Kothavade RJ, Kura MM, Valand AG, Panthaki MH (2010) Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J Med Microbiol 59: 873-880.
- Michael DM, Michael WW, David TC, Catherine RK, Denis JS, et al. (1993) Design of 5-(3, 5-di-tert-butyl-4-hydroxyphenyl)-1, 3, 4-thiadiazoles,-1, 3, 4-oxadiazoles, and-1, 2, 4-triazoles as orally active, nonulcerogenic antiinflammatory agents. J Med Chem 36: 1090-1099.
- Turan-Zitouni G, Kaplancikli Z, Erol K, Kiliç FS (1999) Synthesis and analgesic activity of some triazoles and triazolothiadiazines. Farmaco 54: 218-223.
- Salgin-Gokşen U, Gökhan-Kelekçi N, Göktaş O, Köysal Y, Kiliç E, et al. (2007) 1-Acylthiosemicarbazides, 1, 2, 4-triazole-5 (4< i> H</i>)-thiones, 1, 3, 4-thiadiazoles and hydrazones containing 5-methyl-2-benzoxazolinones: Synthesis, analgesic-anti-inflammatory and antimicrobial activities. Bioorg Med Chem 15: 5738-5751.
- Marzano C, Pellei M, Colavito D, Alidori S, Lobbia GG, et al. (2006) Synthesis, characterization, and in vitro antitumor properties of tris (hydroxymethyl) phosphine copper (I) complexes containing the new bis (1, 2, 4-triazol-1-yl) acetate ligand. J Med Chem 49: 7317-7324.
- Marzano C, Pellei M, Colavito D, Alidori S, Lobbia GG, et al. (2006) Synthesis, characterization, and in vitro antitumor properties of tris (hydroxymethyl) phosphine copper (I) complexes containing the new bis (1, 2, 4-triazol-1-yl) acetate ligand. J Med Chem 49: 7317-7324.
- Holla BS, Veerendra B, Shivananda MK, Poojary B (2003) Synthesis characterization and anticancer activity studies on some Mannich bases derived from 1, 2, 4-triazoles.Eur J Med Chem.38: 759-767.
- Khan I, Ali S, Hameed S, Rama NH, Hussain MT, et al. (2010) Synthesis, antioxidant activities and urease inhibition of some new 1, 2, 4-triazole and 1, 3, 4-thiadiazole derivatives.Eur J Med Chem45: 5200-5207.
- Chohan ZH, Sumrra SH, Youssoufi MH, Hadda TB (2010) Metal based biologically active compounds: Design, synthesis, and antibacterial/antifungal/cytotoxic properties of triazole-derived Schiff bases and their oxovanadium (IV) complexes. Eur J Med Chem 45: 2739-2747.
- Aher NG, VS Pore, Mishra NN, Kumar A, Shukla PK, et al. (2009) Synthesis and antifungal activity of 1, 2, 3-triazole containing fluconazole analogues. Bioorganic Med Chem Lett 19: 759-763.
- Camoutsis C, Geronikaki A, Ciric A, Soković M, Zoumpoulakis P (2010) Sulfonamide-1, 2, 4-thiadiazole derivatives as antifungal and antibacterial agents: synthesis, biological evaluation, lipophilicity, and conformational studies.Chem Pharm Bull58: 160-167.
- Zoumpoulakis P, Camoutsis C, Pairas G, Soković M, GlamoÄÂlija J, et al. (2012) Synthesis of novel sulfonamide-1, 2, 4-triazoles, 1, 3, 4-thiadiazoles and 1, 3, 4-oxadiazoles, as potential antibacterial and antifungal agents. Biological evaluation and conformational analysis studies.Bioorg Med Chem 20: 1569-1583.
- Sangshetti JN, Lokwani DK, Sarkate AP, Shinde DB (2011) Synthesis, Antifungal Activity, and Docking Study of Some New 1, 2, 4â€Âtriazole Analogs. Chem Biol Drug Des 78: 800-809.
- Meis JF, Verweij E (2001) Current management of fungal infections. Drugs 61: 13-25.
- Muhammad SA, Ali A, Ismail T, Zafar R, Ilyas U, et al. (2014) Insilico study of anti-carcinogenic lysyl oxidase-like 2 inhibitors. Comput Biol Chem 51: 71-82.
- Odds FC, Brown AJ, Gow NA (2003) Antifungal agents: mechanisms of action.Trends Microbiol 11: 272-279.
- Muhammad SA, Ali A, Ismail T, Zafar R, Ilyas U, et al. (2014) Insilico study of anti-carcinogenic lysyl oxidase-like 2 inhibitors. Comput Biol Chem 51: 71-82.
- Lagorce D, Sperandio O, Baell JB, Miteva MA, Villoutreix BO (2015) FAF-Drugs3: a web server for compound property calculation and chemical library design.Nucleic Acids Res 43: 200-207.
- Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1: 337-341.
- Pollastri MP (2010) Overview on the Rule of Five. Curr Protoc 9: 2.
- Hassan M, Ashraf Z, Abbas Q, Raza H, Seo SY (2016) Exploration of Novel Human Tyrosinase Inhibitors by Molecular Modeling, Docking and Simulation Studies.Interdiscip Sci: 1-13.
- Dunkel M, Günther S, Ahmed J, Wittig J, Preissner R (2008) SuperPred: Drug classification and target prediction. Nucleic Acids Res 36: 55-59.
- Hooft RW, C Sander, Vriend G (1997) Objectively judging the quality of a protein structure from a Ramachandran plot. Comput Appl Biosci 13: 425-430.
- Yang J, Roy A, Zhang Y (2013) Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinfo 29: 2588-2595.
- Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263: 243-250
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi