Review Article, J Appl Bioinformat Computat Biol Vol: 0 Issue: 0
Assessment And Application of EEG: A Literature Review
John Reaves1, Timothy Flavin1, Bhaskar Mitra2, Mahantesh K3 and Vidhyashree Nagaraju1*
1Tandy School of Computer Science, The University of Tulsa, Tulsa, OK 74104 USA
2Idaho National Laboratory, Idaho Falls, ID, USA
3Department of Electronics & Comunication Engineering, SJB Institute of Technology, Bengaluru, KA, India
*Corresponding Author: Vidhyashree Nagaraju
Tandy School of Computer Science, The University of Tulsa, Tulsa, OK 74104 USA
E-mail: vidhyashreenagaraju@utulsa.edu
Received: July 01, 2021 Accepted: July 19, 2021 Published: July 26, 2021
Citation: Reaves J, Flavin T, Mitra B, Mahantesh K, Nagaraju V (2021) Assessment And Application of EEG: A Literature Review. J Appl Bioinforma Comput Biol 10:7.
Abstract
Advancements in neuroscience have enabled the collection and assessment of neurological data to assist in the detection and treatment of several medical conditions as well as the operation and control of devices through brain-computer interface. Existing studies rely heavily on data such as elec- troencephalography (EEG) because of its ease of data collection and assessment, high temporal resolution, low cost, ease of use, and high computational accuracy compared to other neurological and physiological data such as functional Magnetic Resonance Imaging (fMRI) and heart rate.
This paper provides a comprehensive review of recent liter- ature on EEG data assessment and applications in computer and neuroscience research. Specifically, the paper reviews articles recently published in high impact venues including IEEE to provide a brief insight into ongoing work in this research area. The survey is intended to provide a quick summary for researchers, graduate students, and any interested individuals seeking to advance research on this topic. It should also be beneficial to neuroscientists and professionals wishing to obtain a quick overview of previous work. In addition to summarizing key methodologies on data collection, preprocessing, and algorithms, we identify open data sets, software, and developing trends that would benefit from continued exploration.
Keywords: Electroencephalograph, Brain-Computer Interface, Magnetic Resonance Imaging, Data Preprocessing, Feature Extraction, Algorithms, Machine Learning
Introduction
Technological advancement in the field of neuroscience has enabled solutions to many problems through the collection and assessment of neurological and physiological data such as Electroencephalography (EEG) [1], functional Magnetic Resonance Imaging (fMRI) [2], Electrocardiogram (ECG) [3], Electromyography (EMG) [4], and Galvanic Skin Response (GSR) [5]. Specifically neurological signals such as EEG and fMRI have assisted in advancing the detection and treatment of many neurological afflictions, including mental health dis- orders, through neurofeedback signals specific to the patient. Additional areas include improved focus in learning and design of brain-computer-interface (BCI) [6,7] devices, including brain-wavecontrolled prostheses and control of autonomous systems.
Much progress has been made over the years in understanding and applying EEG and other types of data. Other for- mats, such as electrocorticography (ECoG), sometimes called intracranial EEG and fMRI, are gaining traction. However, these methods of data collection have limitations that restrict their usage [8]. A review of existing literature suggests that applications of EEG ranging from emotion discrimination and seizure detection to motor imagery (MI) in BCI have seen significant increases in accuracy and computation speed. How- ever, there are challenges in filtering out noise and acquiring detailed information from the observed signals. Some of the challenges encountered are:
• Limited or lack of communication between neuroscien- tists and computer scientists.
• Lack of availability of recent datasets to support algorithmic research.
• Limited knowledge of the data collection process and application area that may hinder the ability to develop or apply algorithms without losing important information.
• Availability of variety of preprocessing pipelines for use in EEG applications to remove artifacts. While there is no qualityassurance standardization between methods as yet, continuing to encourage diverse approaches while having a means to evaluate them may be preferred to a central “gold standard” [9].
• Inherent non-stationarity of EEG signals complicates generalization, as signals vary between subjects and within subjects, depending on their state of mind.
This paper provides a comprehensive review of articles on EEG approaches published in high-impact journals and conferences including IEEE, Springer, and Elsevier. 48 articles from 15 journals and 3 conferences in IEEE, 22 papers from 17 journals in Springer, and 24 articles from 8 journals in Elsevier were selected from a pool of over 700 articles and analyzed. Much attention was paid to highimpact journals, including IEEE Machine Learning and Pattern Recognition, Elsevier Pattern Recognition, and proceedings including the Genetic and Evolutionary Computation Conference and the International Conference on Bioinformatics and Biomedical Science. The shortlisted articles capture the limitations and challenges of EEG research as well as highlight efforts from computer science researchers. The survey focuses on summarizing data collection, preprocessing, feature extraction, and classification/prediction algorithms. It provides a quick review of the existing and latest work for people who would like to pursue research in this area and enhance our understanding. The survey is intended to support graduate students, academic researchers, neuroscientists, and other professionals in the field. The paper also includes a list of devices and open data sets to support continued exploration.
The remainder of the paper is organized as follows: Section II provides a definition of EEG along with a brief description of data and devices, while Section II-B identifies publicly available datasets. Section III outlines some of the applications of EEG. Section IV reviews different stages of data processing and highlights efficient and commonly used algorithms at each stage. Section V provides a summary of the study along with some directions for future research.
Electroencephalography
This section describes EEG along with its benefits and drawbacks. It then summarizes EEG data collection and visualization with an example, and identifies publicly-available datasets. Finally, invasive and non-invasive data collection de- vices are discussed, and commercially available EEG devices are identified with their technical specifications.
A. Definition and Description
EEG is a measure of electrical activity in the brain that records frequencies observed through the brain’s normal activity. While EEG signals were discovered in 1875 through Richard Caton’s work with animals [9,10], the term ‘EEG’ was coined by Dr. Hans Berger in 1924 after the successful recording of the first human electroencephalograph. Formally, Olejniczak [1] defines EEG as “a graphic representation of the difference in voltage between two different cerebral locations plotted over time,” mostly consisting of synaptic activity, though contaminated with noise from other sources and distorted by being measured through the skull. Initially a novelty, interest in EEG technology increased with the discovery of seizure patterns.
EEG is prominently used in biomedical applications for the detection of neurological disorders such as epilepsy, tumors, sleep disorders, and inflammation or damage in the brain. In addition to this, EEG is extensively used in neuroscience research focused on, but not limited to, motor, cognitive, and sensory imaging. Advances in neuroscience research have enabled the development of braincomputer interfaces, which facilitate the control and use of devices via brain wave interpretation.
EEG is generally preferred over other methods of data collection mainly because of its high temporal resolution, low device cost, noninvasive and easy data-collection process as well as the fact that EEG data conversion and interpretation is computationally less expensive than other methods. However, EEG has a low signal-to-noise ratio since brain activity is observed through the skull, and motion can add additional noise artifacts. As mentioned before, signals are not always consistent between or within individuals. While individual differences are beneficial in applications such as EEG-based biometric identification, it complicates any kind of generalizable BCI or other algorithms that attempt to understand the functional details of the brain. This complication is further worsened by differences in individuals’ brain waves at any given time, based on emotional state, movement, and so on.
B. Data Collection and Interpretation
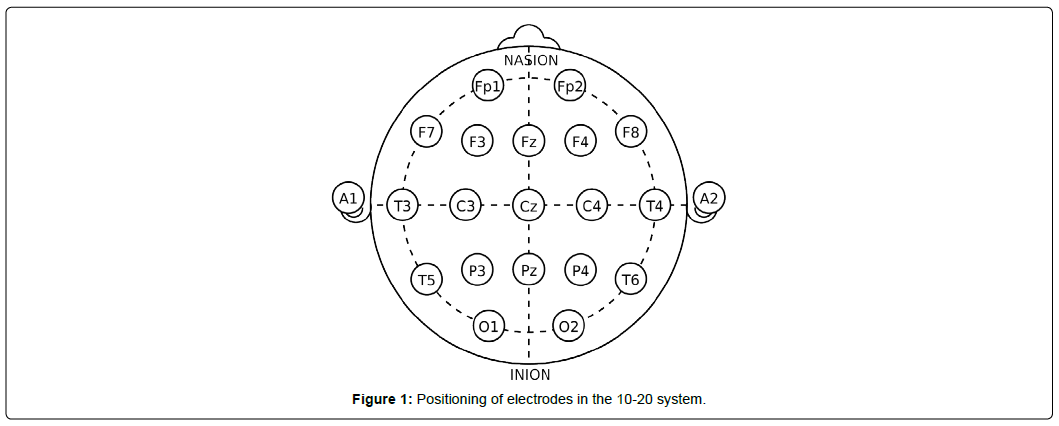
The EEG data collection process is typically centered around particular frequencies depending on the specific ap- plication, such as a research problem or medical assessment. Collection of EEG data through electrode placement adheres to internationally agreed rules [11], generally classified into 10-10 or 10-20. The numbers refer to the distance between electrodes; in the 10-20 system, for instance, electrodes are 10% of the skull’s left-right distance and 20% of its front-back distance apart. The placement starts with initial marks at four points: between the forehead and nose, middle of the back of the skull over the occipital area, and on both sides of the head above the outer part of the ear opening. After the indentation, the electrodes are placed at specific distances from the points. The brain signals can be localized by narrowing down the region through the addition of electrodes.
Figure 1 shows electrode placement based on 10-20 system to collect EEG data.
Figure 1 shows connection points for 21 total channels, where each channel corresponds to an electrode and outputs a waveform. The connection points or electrodes are denoted by letters and numbers to easily distinguish them. The letters correspond to lobes, or approximate parts of the brain being analyzed: frontopolar or prefrontal cortex (Fp), frontal (F), temporal (T), parietal lobe (P), occipital (O), auricular (A), and central (C). The ‘Z’ label associated with these letters indicates electrodes along the midline of the head. The left and right hemisphere of the brain are identified by odd and even numbers, respectively.
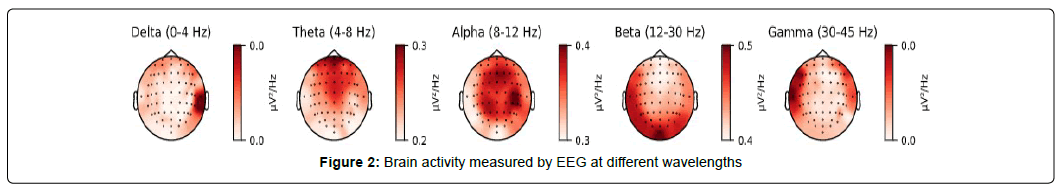
Common wavelengths used in EEG analysis include delta (<4 Hz), theta (4-8 Hz), alpha (8-12 Hz), beta (12-30 Hz), and gamma (30-45 Hz) waves as shown in Figure 2. Figure 2 shows a visualization of brain activity [12,13] at different wave- lengths generated using MNE [14] Python.
Alpha and beta waves in Figure 2 are commonly used in motor imagery applications, with the former correlating with eye-muscle movements and the latter associated with general movement [10]. In emotion and preference research, alpha waves are associated with positive emotions while asymmetrical, and greater beta waves (16-18 Hz) are tied to individual preference. Higher and lower frequency in theta bands indicates positive and negative emotions, respectively [15]. Delta waves in Figure 2 are assumed to be less useful and filtered out during the process of noise reduction.
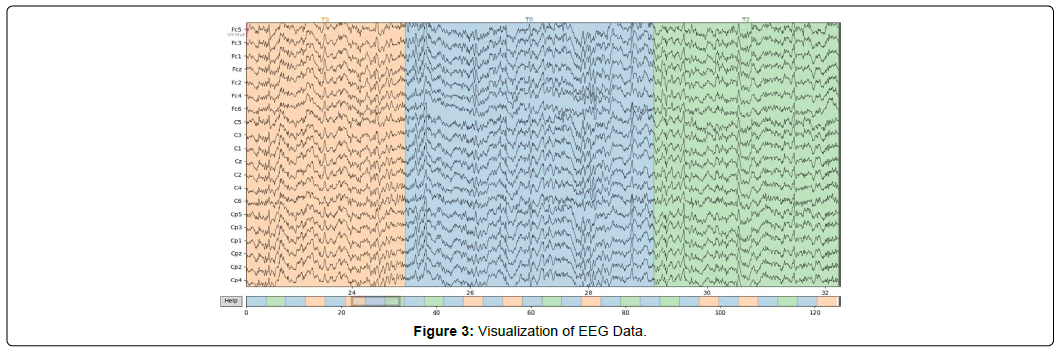
3 is a visualization of an EEG motor movement/imagery dataset [13] collected using non-invasive device described in Figure 1 and Figure 2. In Figure 3, the x-axis indicates time in seconds and the y- axis corresponds to the output of each electrode described in Figure 1, sampled at 160 times per second. Figure 3 presents unprocessed data with no visible effect of the artifacts, which are commonly introduced during EEG data collection. The color changes represent the transitions between different events, with an approximate length of 4s along with the resting state (T0) between events. In baseline trials T0 is recorded between 60-120s.
Additional data formats: Other formats considered as standalone replacements or multimodal supplements in neuro- science research and development include:
• ECoG: This method is similar to an invasive form of an EEG scan. Subdural electrodes measure activity directly from the surface of the cerebral cortex, but the invasive nature of data collection limits its adoption [8].
• fMRI: This approach measures activity more spatially through detection of blood oxygen level-dependent changes in the MRI signal due to neuronal activities as a result of a stimulus or task [2]. While useful, the amount of time required to take a clear picture and potential for noise introduction severely limit the feasibility of its real- time use.
• EMG: An EMG analyzes electrical activity in the muscles. EMG is useful in examining the connection between nerves and muscles in a particular part of the body. Due to spatial limitations, EMG is often used for diagnosis rather than for signal analysis.
In addition to these formats, eye movement, heart rate, and body temperature are sometimes used in combination with EEG and other data formats to improve the accuracy of classification or prediction.
Publicly Available Datasets
Table 1 summarizes some publicly and readily available datasets for EEG that should serve as a helpful resource for subsequent work in the field.
| Name | Data type | Reference |
|---|---|---|
| OpenNeuro | EEG | [16] |
| BCI Competition | EEG | [17] |
| Physionet | EEG | [18], [19] |
| UCI-ML | EEG | [20], [21] |
| EEGLab | EEG/ERP | [22] |
| DEAP | EEG | [23] |
| BNCI Horizon 2020 | EEG/EMG/EOG | [24] |
| EEGbase | EEG/ERP | [25] |
Table 1: Publicly available EEG data repository.
In Table 1, the BCI Competition datasets [17] and DEAP [18-24] are among the most commonly used for MI activities and emotional interpretation, respectively. In addition, Temple University maintains a comprehensive list of EEG databases and complementing software. Additional neurological data sets are maintained by Neuroscience Information Framework [25-27], while MRI and fMRI sources are maintained by Neuro- data [28] and OpenfMRI [29], respectively.
C. Devices
EEG data is typically collected by placing electrodes on the skull similar to Figure 1. At a high level, devices used to collect EEG data can be classified into invasive and non- invasive depending on placement of electrode inside or outside the skull.
1. Non-invasive: Information gathered from the brain is gathered without any surgical procedure. The data is usually collected using a cap or headset.
2. Invasive: Data is gathered from electrodes placed directly on the brain. Collected data will be less susceptible to noise and other interference. Additional benefits include accurate localization of signals.
The non-invasive device is the preferred type of data collection due to its ease of use and low cost. While selecting a non- invasive device, there are additional parameters that should be considered such as:
• Electrode Patterns: Selection of patterns including the original 10-20 or 10-10 system. Different and denser patterns usually allow for greater level of detail [30].
• Channels: The quality of data collection and assessment is dependent on electrode placement and the number of channels analyzed. Modern devices typically feature 24- 32 and up to 128 channels [31]. While a greater number of channels can allow for greater level of detail, it also increases the setup time and device cost significantly. However, the cost remains less expensive compared to fMRI data collection.
• Sensor Technology: EEG electrodes can be wet, dry, or semidry. Dry electrodes are easier to set up [32], but can be prone to interference and motion artifacts, unlike wet technology, which uses a conductive cream or paste. Semi-dry technology uses polymer electrolyte, which is usually utilized in devices such as EEG buds to record data for long periods of time.
• Connection: can be wired or wireless. Wireless connection usually involves transmission of data over Bluetooth or similar technology. While wireless is more expensive, such a feature is convenient in a laboratory setting, especially if movement of any sort is involved.
• Device type: could be a cap, headset, headband, or buds depending on the type of use. Caps and headsets are most commonly used in laboratory settings to collect high- quality data, whereas headbands and buds are mostly used in applications involving cognitive studies.
Table 2 lists commercially-available EEG devices and their approximate cost. Many devices feature accompanying soft- ware to process, visualize, and analyze the data, also listed in Table 2.
| Device Name | Data type | Channels | Device Style | Electrode Type | Data Transfer | Freq. Range | Software | Cost | Ref |
|---|---|---|---|---|---|---|---|---|---|
| Emotiv EPOC Flex | EEG | ≤ 32 | Cap | Wet | BT | 0.2 - 45Hz | EmotivPRO | ≥$1700 | [33] |
| Emotiv EPOC X | EEG | 14 | Headset | Wet | USB, BT | 0.16 – 43Hz | EmotivPRO | $850 | [34] |
| OpenBCI | EEG | 16-Aug | Cap, Headset | Dry, Wet | Wired, BT | - | OpenBCI (Open-Source) | $1,000 | [35], [36] |
| NeuroSky MindWave | EEG | 1 | Headset | Dry | BT | 3-100Hz | NeuroView, NeuroSkyLab | $100 | [37] |
| Muse 2 | EEG, Multi | 4 | Headset | Dry | BT | 0.5-100Hz | Muse/Muse Direct Apps | $250 | [38] |
| MindMedia NeXus 10 | EEG, Multi | 8 | - | - | - | - | BioTrace+ | $1,000 | [39] |
| ActiveTwo | EEG, Multi | 32 | - | - | USB | None | - | $1,000 | [40] |
Table 2: Devices.
Applications
This section identifies and describes the applications of EEG data in medical and non-medical categories.
A. Medical Applications
With the increased interest in EEG technology following the discovery of epilepsy spikes in 1930, seizure detection has long been a major biomedical application of EEG data. Identification and prediction of seizures [33-42] in epileptic patients has significantly improved the quality of patients’ lives and enhanced the reliability of medical treatments. Ap- plications for epileptics are getting faster and more cost- effective as researchers continue to uncover new algorithms with high detection [43] and prediction accuracy [41] as well as improved epileptogenic foci localization [44].
Motor imagery [45] is another prominent field that attempts to understand and map human thinking processes into action. MI research is essentially predicated off the fact that whether someone actually moves a limb or simply imagines moving a limb, the brain signals produced are the same. Essentially, a functioning MI program could allow amputees [46] to regain movement in robotic versions of their lost limbs or allow anyone to remotely operate such limbs. The present state of the field is limited and classification of brainwaves is restricted to a few degrees of freedom. Recent studies on robotic prostheses focus on control [47], while some studies have focused on achieving that control with a short training time of approximately 15 minutes [48]. In addition, to improve the classification of signals, physiological data such as facial expression can also be analyzed [49].
Another major application of EEG is in the assessment and development of rehabilitation methods. Recently, a real-time EEG based MI-BCI system with a virtual reality game [50] was developed as a motivational tool with feedback for patients in stroke rehabilitation. Another study [51] explored the possibility of using por personal EEG devices with games to motivate patients to carry out their rehabilitation exercises. Additional applications include individual preference identification through interpretation of emotional information, especially for those with difficulties communicating [52,53] the study of sleep disorders through sleep-quality assessment [54] the analysis of EEG signals during pain perception [55]; the classification of potentially alcoholic patients [56] and the diagnosis of Parkinson’s disease [57].
B. Non-medical Applications
Non-medical applications of EEG are focused on, but not limited to, cognitive studies, robotic prostheses and security systems. Previous research on cognitive studies involves emotion recognition [52] and classification [23]. In addition, physiological signals were combined with EEG to improve the accuracy of emotion recognition [58]. A few studies also focused on methods to improve focus in e-learning through. attention feedback [59], improve learning for novice programmers through neurological signals-controlled interface [59,60] and to report experiences [61] of using EEG in the context of a software engineering education.
While robotic prostheses have significant use in medical applications, computer researchers are focused on utilizing motor imagery in applications including autonomous system operation and control [62,63]communication, and security research [64]. Recent studies include one focused on the tele- operation of a dualarm robot carrying a common object in multi-fingered hands [65]. The study is then extended to a controllable multi-directional arm to reach tasks in three- dimensional environments [7]. Many studies have also improved the classification accuracy of motor-imagery signals [66]. EEG signals are also used in security, specifically in biometrics authentication systems. EEG signals are used for personal identification [67] including facial recognition [68]. Most recently, one such study [69] examined network patterns and graph features to understand the distinctiveness of humans’ EEG functional connectivity and provide useful guidance for the design of graphbased EEG biometric systems.
EEG signals are also used in vigilance detection [70,71] critical for those who engage in long, demanding tasks such as monitoring systems or driving, which makes it a key field in BCI research. Additional non-medical applications include speech recognition [72] user intention classification [73] driver fatigue evaluation [47] for traffic safety, and mental workload assessment [74] for maintaining mental health and preventing accidents. More unique studied examples include using EEG as a lie detector [75] and using it for evaluating one’s confidence in making decisions [76].
Data Processing
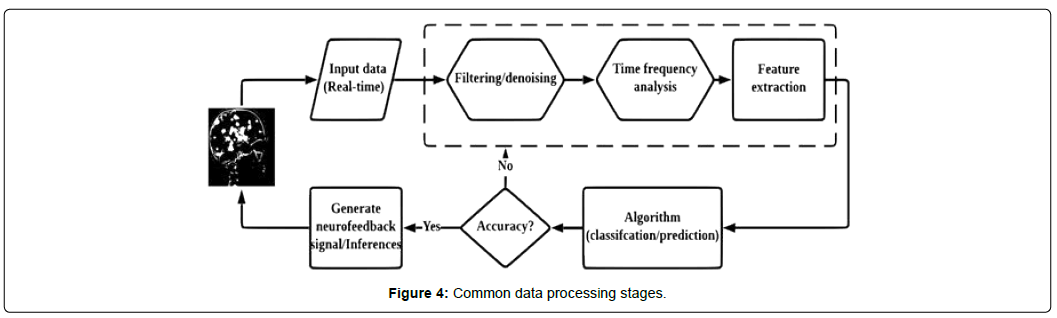
This section describes steps in EEG data processing, including the preprocessing and classification stages. The section also summarizes existing studies on each stage of EEG data processing and identifies corresponding machine learning algorithms [77,78]. Figure 4 outlines the stages of EEG data processing in a neurofeedback system. In Figure 4, stages identified in the dashed box corresponds to data preprocessing. Most of the existing studies detail one or two stages of preprocessing rather than all three, due to either lack of information on the collection process or lack of knowledge in the selection and application of preprocessing methods.
A. Denoising
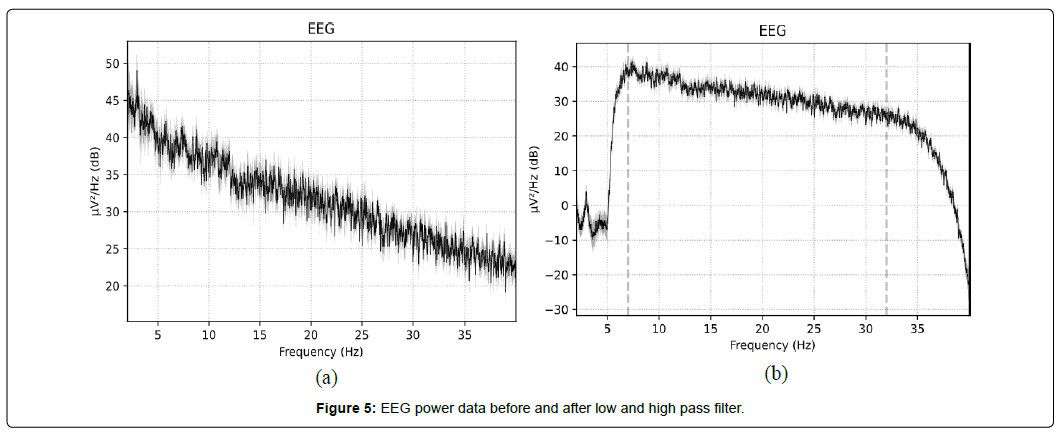
In reducing and removing noise, the most common approaches include using low, high, or bandpass filters. These filters allow the user to pass along frequencies above, below, or between specified values, respectively. The choice of filters and frequency selection is dictated by the specific task at hand; for example, waves above 30Hz and below 8Hz will often be filtered out in studies involving motor imagery, as they are less relevant. More generally, very low frequencies are often muscle-movement artifacts, while frequencies in the 50- 60 Hz range feature noise from power lines or other electronic signals. There are a variety of more dynamic filters in use, such as Volterra [79] the zerophase bandpass Butterworth filter [80,81] and even fully adaptive filters for epilepsy detection [82,83].
Figure 5 shows a visualization of power recorded in the EEG data in Figure 3 after passing through low- and high- pass filters.
While denoising using filters enables significant noise reduction by selecting only the required frequency range, artifacts in the data are more difficult to remove. Therefore, many studies extend the denoising stage with methods such as Independent Component Analysis (ICA) in attempts to automatically remove such artifacts [84- 86] though earlier and modern studies sometimes resort to manual artifact removal [87]. Infrequently, researchers and practitioners use filtered data directly without applying artifact removal procedures, though this has caused significant deviations in the overall assessment.
B. Time-frequency
Considering the non-stationary nature of EEG data, timefrequency analysis provides a temporal indication of multiple artifacts with the feature of time, helpful for developing controllers and feedback devices. Artifact removal [88] is a critical, yet challenging step in processing EEG data. The challenge is to analyze and isolate the data and remove the influence of other activities, such as prominent eye blink, heart rate, facial movement, and body temperature. Existing studies treat artifact removal as an optional stage. Less than 50% of the studies discuss artifact removal, with over 80% focused on manual removal based on their domain and problem-specific knowledge ICA and discrete wavelet transform (DWT) are among the most commonly used approaches to remove artifacts. However, application of these approaches are limited to static data analysis in applications such as epileptic seizure detection. However, some of these studies have highlighted the use of time-frequency analysis in reducing artifacts by assessing the input signal in both the time and frequency domains to achieve better resolution. For EEG data in particular, wavelet transforms (WT) [81] are commonly employed to more easily work with the inherently non-stationary signal. While WT can also help with denoising, it requires a relatively involved reconstruction process that is not used when using simpler filters; this process in itself can aid in dimensionality reduction, making the information that remains significantly more useful. As such, the Flexible Analytic Wavelet Transform (FAWT) [88,89] and others are extremely common due to their relative effectiveness. Common Spatial Patterns (CSP) [45, 80, 90] are also frequently used to preserve the limited spatial data ascertainable from an EEG, though the computational time that this takes limits its usefulness in real-time applications. Because of the extent to which the data can be altered by these processes, there are a few ways to estimate the relative quality of data afterwards. These include Harvard’s HAPPE, the LTAPP, and Automagic [91]. Considering how important the quality of the data used to develop a model is, these tools may see wider future use.
Additional time-frequency analysis methods include the Fourier transform (FT), short-time Fourier transform (STFT), Hilbert Huang transform (HHT), etc. FT is used only to deconstruct the received signal into component frequencies, though time information is necessarily lost. Because of the non-stationary nature of EEG signals, this is an uncommon approach and it is mostly coupled with other methods when used. Santoso [92] describes STFT as an extension of FT, which is able to preserve time information through the use of windowing, where FT will be applied to the subset of data in each window. A review of existing studies suggests the broad utilization of WT, which is able to preserve both time and frequency information. Specifically, WT is good in the time resolution of high frequencies, while for slowly varying functions, the frequency resolution is remarkable. On the more complex end, some authors make use of the HHT, which decomposes a signal into intrinsic mode functions (IMF) that also preserve temporal and spatial information [93]. It is highly effective for non-stationary and nonlinear data like EEG, and is complex enough that it could be considered an algorithm rather than a particular tool.
C. Feature extraction
Most commonly, regression and similar machine learning methods [94, 95] are used for feature extraction. Some of the most commonly used methods include logistic regression [42] support vector machine (SVM) [71] linear discriminant analysis (LDA) [96], principal component analysis (PCA) [97], evolutionary algorithms [98], and ensemble learning [99] including random forest [94] and XgBoost learning [94].
Recent studies have employed neural networks (NN) as the most common method of feature selection because of their ability to process information despite noise or artifacts. Convolutional [100] and analytical [101] neural networks with some variations, such as recurrent neural networks with long short-term memory (LSTM) are most commonly used. CSP algorithms are also frequently seen [45,80,90] especially in combination with methods of denoising that already decompose the signals. On the opposite end of the spectrum, a few papers [102,103] make use of fusional features, attempting to combine information from different electrodes, frequencies, or both in order to reduce the number of dimensions analyzed. This makes computation simpler and faster but runs the risk of reducing accuracy.
Less-common methods include recurrence quantification analysis (RQA) [104] a quadratic time-frequency distribution (QTFD) and Choi- Williams distribution (CWD) [105] quadratic discriminant analysis [106] to detect changes be- tween states, various types of segmenting [107, 108] and even the NSGA-II genetic algorithm [98]. Many studies, including [98] combine multiple methods of feature selection and machine learning, often using neural networks or k- nearest neighbors in tandem with more complex or uncommon methods.
Preprocessing algorithms: Table 3 lists most commonly- used algorithms and corresponding studies in EEG preprocessing.
| Algorithm | Reference |
|---|---|
| Wavelet transform (II) | [42]–[44], [70], [74], [81], [84], [86], [87], [89], [96], [97], [100], [101], [109]–[120] |
| Fourier transform (II) | [53], [86], [87], [92], [99], [120] |
| Independent component analysis (II-III) | [7], [57], [68], [69], [83]–[87], [91], [96], [119]–[123] |
| Principal component analysis (II-III) | [47], [58], [83], [89], [97], [120], [121], [124]–[126] |
| Power spectral density (I-II) | [81], [85], [126]–[128] |
| Common average reference (I) | [7], [102], [110], [120] |
| Common spatial pattern (II-III) | [45], [65], [80], [82], [90], [93], [99], [120], [121], [128]–[138] |
| Hjorth parameters (I) | [75], [87], [112], [123], [139] |
| Convolutional neural network (III) | [99], [117], [129], [140], [141] |
| Discrete cosine transform (II) | [86], [99] |
| Long short-term memory (III) | [56], [142]–[144] |
| Linear discriminant analysis (II-III) | [96], [128] |
| Genetic algorithms (III) | [57], [98] |
| Support vector machine (III) | [71], [109] |
| Transfer learning (III) | [145], [146] |
| Empirical mode decomposition (I) | [54], [87], [99] |
| Finite impulse response filter (I) | [82], [147], [148] |
| Adaptive auto regression (I) | [87], [120] |
| Autoregression (III) | [120], [149] |
| Detrended fluctuation analysis (II) | [54], [109] |
| Partial directed coherence (III) | [57], [149] |
| Quadratic time-frequency distribution (II) | [105], [141] |
| Renyi Entropy (III) | [119], [150] |
| Symmetric and positive definite matrix (III) | [151], [152] |
| Least mean squares (II) | [83], [86] |
| Multiple artifact rejection algorithm (I) | [69], [91] |
Table3: Preprocessing.
Algorithms that correspond to different stages of preprocessing are identified as I, II, III to represent denoising, time- frequency analysis, and feature extraction, respectively. While the wavelet and Fourier transforms as well as CSP are the most commonly used techniques in time-frequency analysis since they preserve spatial information, ICA and PCA seem to be the preferred feature extraction approaches. Neural network algorithms along with transfer learning approaches are gaining attention.
D. Classification and Prediction
The classification/prediction stage of data processing is the final stage of EEG data that precedes inferences. The accuracy and computation time of classification is dependent on the quality and number of features extracted during preprocessing. While smaller sample sizes are easier to compute, many algorithms will not be able to make accurate predictions with small training sets. To address this, recent research has focused on applications of machine learning algorithms such as neural networks, which enable transfer and active learning to allow for the utilization of previously trained knowledge while making inferences based on small samples or less studied area.
Table 4 lists most commonly used algorithms and the corresponding studies in classification and prediction.
| Algorithm | Reference | Field | Hybrid |
|---|---|---|---|
| Support vector machine | [23], [42], [44], [45], [47], [48], [53], [55], [56], [58], [65], [66], [68], [80], [86], [87], [93], [96]–[99], [103], [105], [111], [113], [120], [121], [126], [127], [129], [130], [134], [137], [148], [149], [152]–[156] | MI-BCI, Emotion, Epilepsy, Cognitive | Ensemble Group, Sparse |
| k-Nearest neighbor | [42], [53], [55], [58], [68], [96]–[98], [103], [113], [120], [123], [126], [127], [129], [148] | MI-BCI, Emotion, Epilepsy, Cognitive | CFS+KNN |
| Neural network | [41], [45], [54], [73], [87], [97], [98], [101], [117], [118], [120], [147] | MI-BCI, Epilepsy, Cognitive, Sleep | DNN, ANN, MLP |
| Convolutional NN | [7], [41], [50], [56], [74], [81], [92], [100], [110], [116], [117], [132], [137]–[139], [141], [157]–[165] | MI-BCI, Emotion, Cognitive, Epilepsy | S-EEGNet, R3DCNN, CNN-LSTM |
| Linear discriminant analysis | [48], [58], [82], [87], [89], [93], [98], [113], [120], [121], [129], [131], [135]–[137], [149], [155] | MI-BCI, Sleep | KLDA, LDA, Shrunken |
| Long Short Term Memory | [7], [41], [52], [138], [159], [166]–[168] | MI-BCI, Emotion, Epilepsy, Cognitive | CNN-LSTM |
| Random Forest | [42], [53], [57], [68], [72], [103], [111], [127] | Emotion, Cognitive, Epilepsy | - |
| Naive Bayes | [23], [53], [68], [97], [120], [148] | Emotion, Sleep, Cognitive | Gaussian |
| Recurrent NN | [41], [42], [164], [169] | MI, BCI, Epilepsy, Biometric | R-3DCNN |
| Decision Tree | [42], [58], [68], [97], [113] | MI-BCI, Emotion Epilepsy, Sleep | - |
| Transfer Learning | [127], [138], [145], [146], [170] | MI-BCI, Emotion | DTL, MFTL, CSP, DNN |
| Logistic Regression | [42], [58], [126], [129] | MI-BCI, Emotion, | |
| Common spatial pattern | [155], [170], [171] | MI-BCI | Filter Bank, Sparse Filter Band |
| Deconvolutional NN | [47], [127] | MI-BCI, Emotion | - |
| Clustering | [43], [87] | Epilepsy, Sleep | K-medoids |
| Quadratic discriminant analysis | [106], [113] | MI-BCI | - |
| Principal component analysis | [152], [155] | MI-BCI | KPCA |
| Sparse Representation | [70], [171] | MI-BCI, Cognitive | - |
Table4: Classification
The first column in Table 4 suggests that the first few entries are the most commonly used algorithms in classification. While the popularity of SVM is less clear, the application of neural networks is a promising path towards knowledge- sharing and improved accuracy. In addition, the use of transfer learning as a tool appears more often in classification efforts than in preprocessing. Given the inherent differences in EEG signals within and between people, such an approach will be invaluable in shortening learning times and improving future systems. Transfer learning’s lack of current popularity should not be confused for a lack of importance.
Conclusions and Future Research
This paper presents a comprehensive review of recent research on EEG data processing. The survey summarizes articles shortlisted from high-impact journals and conferences published in venues including IEEE, Elsevier, and Springer. The paper also provides descriptions of EEG data, applications, devices, and the various data processing stages. Comprehensive lists of publicly-available datasets, commercially- available devices, and algorithms that correspond to different data processing stages are provided for researchers and practitioners interested in advancing the field.
Future research will include the examination of commonlyapplied algorithms identified in the article and assess their suitability for EEG data. The development of efficient and generalizable preprocessing approaches that retain temporal and spatial resolution will be considered [109-171].
Acronyms
ANN Artificial neural network
BCI Brain-computer interface
BT Bluetooth
CFS Correlation-based feature selection
CNN Convolutional neural network
CSP Common spatial pattern
CWD Choi-Williams Distribution
DNN Deep neural network
DTL Deep transfer learning
DWT Discrete wavelet transform
ECG Electrocardiography
ECoG Electrocorticography
EEG Electroencephalograph
EMG Electromyography
EOG Electrooculography
ERP Event-related potential
FAWT Flexible analytic wavelet transform
fMRI Functional magnetic resonance imaging
T Fourier transform
GSR Galvanic skin response
HHT Hilbert-Huang transform
ICA Independent component analysis
IMF Intrinsic mode function
KLDA Kernel LDA
LDA Linear discriminant analysis
LSTM Long short-term memory
MI Motor imagery
MLP Multilayer perceptron
MRI Magnetic resonance imaging
NN Neural network
PCA Principal component analysis
QTFD Quadratic time-frequency distribution
RQA Recurrence quantification analysis
STFT Short-time Fourier transform
SVM Support vector machine
WT Wavelet transform
References
- Olejniczak P (2006) Neurophysiologic Basis of EEG. J Clini Neurophysiol 23:186–189.
- Gore J (2003) Principles and Practice of Functional MRI of the Human Brain. The J of clin investigation 112: 4–9.
- Geselowitz D (1989) On the Theory of the Electrocardiogram. IEEE 77:857–876.
- Mills K (2005) The Basics of Electromyography,” J Neurol Neurosurg Psychiatry76: 32-35.
- Shi Y, Ruiz N, Taib R, Choi E, Chen F (2007) Galvanic Skin Response (GSR) as an Index of Cognitive Load,” in Extended Abstracts on Human Factors in Computing Systems 2651–2656.
- Agarwal M, Venkateswaran S, Sivakumar R (2020) Human-in-the- Loop RL With an EEG Wearable Headset: On Effective Use of Brainwaves To Accelerate Learning. ACM Workshop on Wearable Systems and Applications 25–30.
- Jeong, Shim K (2020) Brain-Controlled Robotic Arm System Based on Multi-Directional CNN-BiLSTM Network Using EEG Signals. IEEE Transactions on Neural Systems and Rehabili- tation Engineering 28: 1226–1238.
- Buzs G, a´ki C, Anastassiou A, Koch C (2012) The Origin of Extra- cellular Fields and Currents—EEG, ECoG, LFP and Spikes. Nature Publishing Group Nat Rev Neurosci. 13:407–420.
- Robbins K, Touryan J, Mullen T, Kothe C (2020) How Sensitive Are EEG Results to Preprocessing Methods: A Benchmarking Study. IEEE Trans Neural Syst Rehabil Eng 28: 1081–1090.
- Hosseini M, Hosseini A, Ahi K (2020) A Review on Machine Learning for EEG Signal Processing in Bioengineering. IEEE Reviews in Biomedical Engineering 99:1
- Oostenveld R, Praamstra P (2001) The five percent electrode system for high-resolution EEG and ERP measurements.Clin Neurophysiol 12:713–719.
- Electrode locations of international 10-20 system for eeg (electroencephalography) recording (2001) Retrived from https://commons.wikimedia.org/ wiki/File:21-electrodes-of-International-10-20-system-for-EEG.svg.
- Schalk G, McFarland D, Hinterberger T, Birbaumer N, Wol- paw J (2004) BCI2000: A General-Purpose Brain-Computer Interface (BCI) System. IEEE Transactions on Biomedical Engineering, 51: 1034–1043.
- Gramfort A, Luessi M, Larson E, Engemann D (2003) MEG and EEG Data Analysis With MNE-Python. Frontiers in Neuroscience 7: 267.
- Aldayel M, Ykhlef M, Al A (2020) Electroencephalogram-Based Preference Prediction Using Deep Transfer Learning. IEEE Access 8:176 818–176 829.
- OpenNeuro Public Datasets (2020) Retrived from ” https://openneuro.org/public/datasets, 2020.
- BCI Competitions (2020) retrived from http://www.bbci.de/competition/, 2020.
- Physionet: EEG Motor Movement/Imagery Dataset (2020) retrived from https://physionet. org/content/eegmmidb/1.0.0/, 2009.
- Goldberger A, Amaral L, Glass L (2000) PhysioBank, Phys- ioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 101:220.
- Eeg database (2021) retrived from uci machine learning repository https://archive.ics.uci. edu/ml/datasets/EEG+Database.
- UCI KDD (1999) [Internet] retrived from Archive: EEG database,” http://kdd.ics.uci.edu/databases/ eeg/eeg.html.
- Swartz Center for Computational Neuroscience: EEGLAB. [Internet] retrived from https://sccn.ucsd.edu/∼arno/fam2data/publicly available EEG data.html, 2021, Online; accessed 22-Dec-2020.
- Dabas H, Sethi C, Dua C, Dalawat M, Sethia D (2018) Emotion Classification Using EEG Signals. InT Conference on Computer Science and Artificial Intelligence 380–384.
- BNCI Horizon (2020) [Internet] retrived from http://bnci-horizon-2020.eu/database/data-sets, 2020.
- Neuroscience Information Framework (NIF): EEGbase, retrived from https://neuinfo.org/data/source/nif-0000-08190-1/search?q= nif-0000-08190&l=, 2017.
- Temple University EEG Corpus: Electroencephalography (EEG) Resources (2021) retrived from https://www.isip.piconepress.com/projects/tuh eeg/html/ downloads.shtml, 2021, Online; accessed 03-Feb-2021.downloads.shtml, 2021.
- List of Neuroscience Databases (2020) retrived from https://en.wikipedia.org/wiki/List of neuroscience databases, 2020.
- Open Connectome Project (2020) [Internet] retrived from https://neurodata.io/project/ocp/, 2020.
- OpenfMRI (2020) [Internet] retrived from https://legacy.openfmri.org/, 2020.
- Oostenveld R, Praamstra P (2001) The Five Percent Electrode System for High-Resolution EEG and ERP Measurements. Elsevier Clinical Neurophysiology 112:713–719.
- Casson A (2009) Wearable EEG and Beyond,” Springer Biomedical engi- neering letters 9: 53–71.
- Hinrichs H, Scholz M, Baum A (2000) Comparison Between a Wireless Dry Electrode EEG System with a Conventional Wired Wet Electrode EEG System for Clinical Applica- tions. Nature Publishing Group: Scientific Reports 10:1–14.
- Emotiv EPOC flex (2020) [Internet] retrieved from https://www.emotiv.com/epoc-flex/, 2020.
- Emotiv EPOC X (2020) [Internet] retrived from https://www.emotiv.com/epoc-x/, 2020.
- Openbci (2020) [Internet] retrived from https://openbci.com/, 2020.
- OpenBCI(2020) [Internet] retrived from Software https://github.com/OpenBCI, 2020.
- Neurosky Mindwave Mobile 2 (2020) [Internet] retrived from https://store.neurosky.com/pages/ mindwave, 2020.
- Muse 2(2020) [Internet] retrived from https://choosemuse.com/muse-2/, 2020.
- Nexus-10 (2020) [Internet] retrived from mkii https://www.mindmedia.com/en/products/ nexus-10-mkii/, 2020.
- Cortech ActiveTwo (2020) [Internet] retrived from https://cortechsolutions.com/product-category/ eeg-ecg-emg-systems/eeg-ecg-emg-systems-activetwo/, 2020.
- Chen X, Ji J, Ji T (2018) Cost-Sensitive Deep Active Learning for Epileptic Seizure Detection. Int Conference on Bioin- formatics, Comp Biology, and Health Informatics 226–235.
- Aliyu I, Lim Y, Lim C (2009) Epilepsy Detection in EEG Signal Using Recurrent Neural Network .International Conference on Intelligent Systems, Metaheuristics & Swarm Intelligence 2019:50–53.
- Wu M, Wan T, Wan X, Fang Z, Du Y (2009) A New Localization Method for Epileptic Seizure Onset Zones Based on Time-Frequency and Clustering Analysis. Elsevier Pattern Recognition 111:107687.
- Wang D, Ren D, Li K, Feng Y, Ma D (2018) Epileptic Seizure Detection in Long-Term EEG Recordings by Using Wavelet-Based Directed Transfer Function. IEEE Trans on Biomed Engineering, 65: 2591–2599.
- Baig M, Aslam N, Shum H (2020) Filtering Techniques for Channel Selection in Motor Imagery EEG Applications: A Survey. Springer Artificial intelligence review 53:1207–1232.
- U.S. Department of Veteran Affairs: Office of Research & Development (2021) [Internet] retrived from https://www.research.va.gov/topics/prosthetics.cfm, 2021
- Noel T, Snider B (2019) Utilizing Deep Neural Networks for Brain- Computer Interface-Based Prosthesis Control.
- Mishchenko Y, Kaya E. Ozbay H (2018) Developing a Three-To Six-State EEG-Based Brain-Computer Interface for a Vir- tual Robotic Manipulator Control. IEEE Transactions on Biomedical Engineering, 66: 977–987.
- Miskon A (2019) Identification of Raw EEG Signal for Prosthetic Hand Application. Int Conference on Bioinformatics Research and Applications 78–83.
- Kar T (2009) Brain Computer Interface for Neuro-Rehabilitation With Deep Learning. Classification and Virtual Reality Feedback in Augmented Human International Conference 2019: 1–8.
- Ndulue C, Orji R (2019) Driving Persuasive Games With Personal EEG Devices: Strengths and Weaknesses. Conference on User Modeling. Adaptation and Personalization 173–177.
- Yan J, Chen S, Deng S (2019) A EEG-Based Emotion Recognition Model With Rhythm and Time Characteristics. Springer Brain Infor- matics 6:7
- Bairavi K, Sundhara K (2018) EEG Based Emotion Recognition System for Special Children,” in International Conference on Communication Engineering and Technology 1–4.
- Zhang Y, Xing J, Guo C (2018) Sleep Stage Classifi- cation Based on EEG Signal by Using EMD and DFA Algorithm. International Conference on Robotics, Control and Automation Engineering 2018: 156–160.
- Mansoor Z, Ghazanfar M, Anwar S, Alfakeeh A, Alyoubi K (2018) Pain Prediction in Humans Using Human Brain Activity Data. The Web Conference 359–364.
- Fayyaz A, Maqbool M, Saeed M (2019) Classifying Alcoholics and Control Patients Using Deep Learning and Peak Visualization Method. International Conference on Vision, Image and Signal Processing 2019: 1–6.
- De Oliveira A, de Santana MA (2020) Early Diagnosis of Parkinson’s Disease Using EEG, Machine Learning and Partial Directed Coherence. Springer Research on Biomedical Engineering 36: 311–331.
- Doma V, Pirouz M (2020) A Comparative Analysis of Machine Learn- ing Methods for Emotion Recognition Using EEG and Peripheral Physiological Signals. SpringerOpen Journal of Big Data 7:1–21
- Sethi C, Dabas H (2018) EEG-Based Attention Feedback To Improve Focus in E-Learning International Conference on Computer Science and Artificial Intelligence 321–326.
- Crawford C, Gardner C, Gilbert J (2018) Brain-Computer Interface for Novice Programmers. ACM Technical Symposium on Computer Science Education 32–37.
- Zak T, Nazrul M (2021) Neurological and physiological measures to evaluate the usability and user-experience (UX) of information systems: A systematic literature review. Elsevier Computer Science Review 40: 100375.
- Li R. (2018) An Approach for BrainCcontrolled Prostheses Based on a Facial Expression Paradigm Frontiers in Neuroscience 12: 943, 2018.
- Katyal K, Johannes M S (2014) A Collaborative BCI Approach to Autonomous Control of a Prosthetic Limb System. IEEE International Conference on Systems, Man, and Cybernetics 1479–1482.
- Bernal S, Celdr a´n A, Pe´rez G, Barros M, Balasubramaniam S (2021) Security in Brain-Computer Interfaces: State-of-the-Art, Opportuni- ties, and Future Challenges. ACM Computing Surveys 54:1–35.
- Liu Y (2018) Motor- Imagery-Based Teleoperation of a Dual-Arm Robot Performing Ma- nipulation Tasks. IEEE Transactions on Cognitive and Developmental Systems 11:414–424.
- Elkafrawy N, Hegazy D, Fadel S (2020) Proposed Model for Thought- Based Animation Based on Classifying EEG Signals Using Estimated Parameters and Multi-SVM. International Conference on Informa- tion and Education Innovations 117–121.
- Alyasseri Z, Khader A, Al M, Alomari O (2020) Person Identifica- tion Using EEG Channel Selection With Hybrid Flower Pollination Algorithm. Elsevier Pattern Recognition 107393.
- Bellman C. Martin M, MacDonald S, Alomari R, Liscano R (2018) Have We Met Before? Using Consumer-Grade Brain-Computer In- terfaces To Detect Unaware Facial Recognition. ACM Computers in Entertainment 16:1–17.
- Wang M, Hu J, Abbass H (2020) BrainPrint: EEG Biometric Iden- tification Based on Analyzing Brain Connectivity Graphs,” Elsevier Pattern Recognition 107381.
- Yu H (2010) Vigilance Detection Based on Sparse Representation of EEG. Annu Int Conf IEEE Eng Med Biol Soc 2439– 2442.
- Zhang M, Sun J, Wang Q, Liu D (2020) Classification of Vigilance Based on EEG,” in International Conference on Graphics and Signal Processing 76–80.
- Kumar P, Saini R, Roy P, Sahu P, Dogra D (2018) Envisioned Speech Recognition Using EEG Sensors. Springer Personal and Ubiquitous Computing 22: 185–199.
- Lim C, Lee C, Kim Y (2018) A Performance Analysis of User’s Inten- tion Classification From EEG Signal by a Computational Intelligence in BCI. International Conference on Machine Learning and Soft Computing 2018:174–179.
- Zhang P, Wang X, Zhang W, Chen J (2018) Learning Spatial-Spectral- Temporal EEG Features With Recurrent 3D Convolutional Neural Networks for Cross-Task Mental Workload Assessment. IEEE Trans- actions on Neural Systems and Rehabilitation Engineering 27: 31 –42.
- Bablani A, Edla D, Tripathi D, Kuppili V (2009) An Efficient Concealed Information Test: EEG Feature Extraction and Ensemble Classification for Lie Identification. Springer Machine Vision and Applications 30: 813–832.
- Fernandez J, Valeriani D, Cinel C, Sadras N, Ahmadipour P (2020) Confidence Prediction From EEG Recordings in a Multisensory Environment. International Conference on Biomedical Engineering and Technology 20: 269–275.
- Garg A, Mago V (2001) Role of machine learning in medical research: A survey,” Elsevier Computer Science Review 40: 100370.
- Smiti A (2020) When machine learning meets medical world: Current status and future challenges. Elsevier Computer Science Review 37:100280, 2020.
- Wu X, Zhang Y, Wu X (2019) An Improved Noise Elimination Model of EEG Based on Second Order Volterra Filter. International Conference on Digital Signal Processing 60–64.
- Molla M (2020) Discriminative Feature Selection-Based Motor Imagery Classification Using EEG Signal. IEEE Access
- Collazos D, lvarez A (2020) CNN-Based Framework Using Spatial Dropping for Enhanced Inter- pretation of Neural Activity in Motor Imagery Classification. Springer Brain Informatics 7: 1–13.
- Guger C, Ramoser H, Pfurtscheller G (2002) Real-Time EEG Analysis With Subject-Specific Spatial Patterns for a Brain-Computer Interface (BCI). IEEE Transactions on Rehabilitation Engineering 8: 447–456.
- Torse D, Desai V (2016) Design of Adaptive EEG Preprocessing Algo- rithm for Neurofeedback System. IEEE International Conference on Communication and Signal Processing 0392–0395.
- Noorbasha S, Sudha G. Removal of EOG Artifacts and Separa- tion of Different Cerebral Activity Components. Single Channel EEG—An Efficient Approach Combining SSA-ICA With Wavelet Thresholding for BCI Applications. Elsevier Biomedical Signal Pro- cessing and Control 63: 102168.
- Rejer I, Cieszy Ł (2019) Independent Component Annu Int Conf IEEE Eng Med Biol Soc 22:47–62.
- Sarma P (2016) Pre-Processing and Feature Extraction Techniques for EEGbci Applications-A Review of Recent Research. ADBU Journal of Engineering Technology 5: 1.
- Motamedi S, Moshrefi M, Hill M, Hill C, White P (2014) Signal Processing Techniques Applied to Human Sleep EEG Signals—A Review. Elsevier Biomedical Signal Processing and Control 10: 21–33.
- Zotev V (2014) Self- Regulation of Human Brain Activity Using Simultaneous Real-Time fMRI and EEG Neurofeedback. Elsevier NeuroImage 85: 985–995.
- You Y, Chen W, Zhang T (2020) Motor Imagery EEG Classification Based on Flexible Analytic Wavelet Transform. Elsevier Biomedical Signal Processing and Control 62: 102069.
- Ramoser H, Muller J, Pfurtscheller G (2000) Optimal Spatial Filtering of Single Trial EEG During Imagined Hand Movement. IEEE Trans- actions on Rehabilitation Engineering, 8:441–446.
- Pedroni A, Bahreini A, Langer N (2019) Automagic: Standardized Preprocessing of Big EEG Data,” Elsevier Neuroimage 200: 460–473.
- Santoso I, Purnama I (2020) Epileptic EEG Signal Classification Using Convolutional Neural Networks Based on Optimum Window Length and FFT’s Lengt. International Conference on Computer and Communications Management 87–91.
- Gaur P (2019) An Automatic Subject Specific Intrinsic Mode Function Selection for Enhancing Two-Class EEG-Based Motor Imagery-Brain Computer Interface. IEEE Sensors Journal 19: 6938–6947.
- Russell S, Norvig P (2002) Artificial Intelligence: A Modern Approach.
- Bishop C ( 2006) Pattern Recognition and Machine Learning. Springer.
- Veslin E, Dutra M Bevilacqua L (2019) Lower Gamma Band in the Classification of Left and Right Elbow Movement in Real and Imagi- nary Tasks. Springer Journal of the Brazilian Society of Mechanical Sciences and Engineering 41:91.
- Miranda´I, Aranha C, Ladeir M (2019) Classification of EEG Signals Using Genetic Programming for Feature Construction. Genetic and Evolutionary Computation Conference 1275–1283.
- Leon M, Parkkila C, Tidare J, Xiong N, Astrand E (2020) Impact of NSGA-II Objectives on EEG Feature Selection Related to Motor Imagery. Genetic and Evolutionary Computation Conference 1134–1142.
- Taheri S, Ezoji M, Sakhaei S (2020) Convolutional Neural Network Based Features for Motor Imagery EEG Signals Classification in Brain- Computer Interface System. Springer SN Applied Sciences 2:1–12.
- Shi T, Ren L, Cui W (2019) Feature Recognition of Motor Imaging EEG Signals Based on Deep Learning. Springer Personal and Ubiq- uitous Computing 23: 499–510.
- Alyasseri Z, Khadeer A, Al M, Abasi A, Makhadmeh S (2019) The Effects of EEG Feature Extraction Using Multi-Wavelet Decom position for Mental Tasks Classification. International Conference on Information and Communication Technology 139–146.
- Kinney E, Ebied A (2019) Introducing the Joint EEG-Development Inference (JEDI) Model: A Multi-Way, Data Fusion Approach for Estimating Paediatric Developmental Scores via EEG. IEEE Transactions on Neural Systems and Rehabilitation Engineering 27: 348–357.
- Liu C, Li C, Zhao Z (2018) Research on Feature Fusion for Emotion Recognition Based on Discriminative Canonical Correlation Analysis. International Conference on Mathematics and Artificial Intelligence 30–36.
- Mihajlovi V (2019) EEG Spectra vs Recurrence Features in Understanding Cognitive Effort. International Symposium on Wearable Computers 160–165.
- Alazrai R, Alwanni H (2019) EEG-Based BCI System for Decoding Finger Movements Within the Same Hand. Elsevier Neuroscience Letters 698: 113–120.
- Xu D, Feng Y (2018) Real-Time On-Board Recognition of Continuous Locomotion Modes for Amputees With Robotic Transtibial Prostheses. IEEE Transactions on Neural Systems and Rehabilitation Engineering 26: 2015–2025.
- Sarsfield J, Brown D (2019) Segmentation of Exercise Repetitions Enabling Real-Time Patient Analysis and Feedback Using a Single Exemplar. IEEE Transactions on Neural Systems and Rehabilitation Engineering 27: 1004–1019.
- Mu S. n˜oz-Romero A, Gorostiaga C (2020) Informative Variable Identifier: Expanding Interpretability in Feature Selection. Elsevier Pattern Recognition 98: 107077.
- Hekmatmanesh A, Wu H, Motie A, Li M. Combination of Discrete Wavelet Packet Transform With Detrended Fluctuation Analysis Using Customized Mother Wavelet With the Aim of an Imagery-Motor Control Interface for an Exoskeleton. Springer Multimedia Tools and Applications 78: 503–30 522.
- Alwasiti H, Yusoff MZ, Raza K (2020) Motor Imagery Classification for Brain Computer Interface Using Deep Metric Learning. IEEE Access 8: 109 949–109.
- Gupta V, Chopda MD, Pachori R (2018) Cross-Subject Emotion Recognition Using Flexible Analytic Wavelet Transform From EEG Signals. IEEE Sensors Journal 19: 2266–2274.
- Tanveer A, Salman A (2019) Epileptic Seizure Classification Using Gradient Tree Boosting Classifier. International Conference on Biomedical Engineering and Technology 2019:71–77.
- Chaudhary S, Taran S, Bajaj V, Siuly S (2020) A Flexible Analytic Wavelet Transform Based Approach for Motor-Imagery Tasks Classifi- cation in BCI Applications. Elsevier Computer Methods and Programs in Biomedicine 187:105325.
- Saini M, Satija U, Upadhayay M (2020) Wavelet Based Waveform Distortion Measures for Assessment of Denoised EEG Quality With Reference to Noise-Free EEG Signal. IEEE Signal Processing Letters 27:1260–1264.
- Li Y, Liu Y, Cui W, Guo Y, Huang H et al., Epileptic Seizure Detection in EEG Signals Using a Unified Temporal-Spectral Squeeze- and-Excitation Network. IEEE Transactions on Neural Systems and Rehabilitation Engineering 28:782–794.
- Xu B, Zhang L (2018) Wavelet Transform Time-Frequency Image and Convolutional Network-Based Motor Imagery EEG Classification. IEEE Access 7: 6084–6093.
- Zhang Z, Duan F (2019) A Novel Deep Learning Approach With Data Augmentation To Classify Motor Imagery Signals. IEEE Access 7:945– 954
- Tahir I, Qamar U, Abbas H (2019) Classification of EEG Signal by Training Neural Network With Swarm Optimization for Identification of Epilepsy. International Conference on Machine Learning and Computing 197–203.
- Rahman M, Khanam F, Ahmad M, Uddin M (2020) Multiclass EEG signal classification utilizing Re´nyi min-entropy-based feature selection from wavelet packet transformation. Springer Open Brain informatics 7: 1–11.
- Lakshmi M, Prasad T, Prakash C (2014) Survey on EEG Signal Processing Methods. International Journal of Advanced Research in Computer Science and Software Engineering 4: 1.
- Lotte F (2014) A Tutorial on EEG Signal-Processing Techniques for Mental- State Recognition in Brain-Computer Interfaces. Springer Guide to Brain-Computer Music Interfacing, 2014, pp. 133–161.
- Anders P, Muller H, Skjæret N, Vereijken B Baumeister J (2020) The Influence of Motor Tasks and Cut-Off Parameter Selection on Artifact Subspace Reconstruction in EEG Recordings. Springer Medical & Biological Engineering & Computing 58: 2673–2683.
- Hu B. Li X, Sun S (2016) Attention Recognition in EEG- Based Affective Learning Research Using CFS+ KNN Algorithm. IEEE/ACM Transactions on Computational Biology and Bioinformatics 15: 38–45.
- Paulson K, Alfahad O (2018) Identification of Multi-Channel Simulated Auditory Event-Related Potentials Using a Combination of Principal Component Analysis and Kalman Filtering. International Conference on Biomedical Imaging, Signal Processing 18–22.
- Liu Y, Liu T (2020) Smooth Robust Tensor Principal Component Analysis for Compressed Sensing of Dynamic MRI. Elsevier Pattern Recognition 102:107252.
- Zheng W, Zhu J, Lu B (2017) Identifying Stable Patterns Over Time for Emotion Recognition From EEG. IEEE Transactions on Affective Computing.
- Aldayel M, Ykhlef M, Al A (2020) Electroencephalogram-Based Preference Prediction Using Deep Transfer Learning. IEEE Access 8:176 818–176 829.
- Schwarz A, Brandstetter J, Pereira J (2019) Direct Comparison of Supervised and Semi-Supervised Retraining Approaches for Co-Adaptive BCIs. Springer Medical & biological engineering & computing 57: 2347–2357.
- Zhu X (2019) Separated Channel Convolutional Neural Network To Realize the Training Free Motor Imagery BCI Systems. Elsevier Biomedical Signal Processing and Control 49: 396–403.
- Wang L, Zhang C, Hu X (2018) Time-Frequency-Space Range of EEG Selected by NMI for BCIs .International Conference on Bioinformatics and Biomedical Science 46–52.
- Olias J, Mart R (2019) EEG Signal Processing in MI-BCI Applications With Improved Covariance Matrix Estimators. IEEE Transactions on Neural Systems and Rehabilitation Engineering 27:895–904.
- Sakhavi S, Guan C, Yan S (2018) Learning Temporal Information for Brain-Computer Interface Using Convolutional Neural Networks. IEEE Transactions on Neural Networks and Learning Systems 29: 5619–5629.
- Duan L, Li J (2020) Zero-Shot Learning for EEG Classification in Motor Imagery-Based BCI System. IEEE Transactions on Neural Systems and Rehabilitation Engineering 28: 2411–2419.
- Jiao Y, Zhang Y, Chen X, Yin E, Jin et al., Sparse Group Representation Model for Motor Imagery EEG Classification. IEEE Journal of Biomedical and Health Informatics 23: 631–641.
- He H, Wu D (2019) Transfer learning for Brain-Computer interfaces: A Euclidean Space Data Alignment Approach. IEEE Transactions on Biomedical Engineering 67:399–410.
- Netzer E, Frid A, Feldman D (2020) Real-Time EEG Classification via Coresets for BCI Applications. Elsevier Engineering Applications of Artificial Intelligence 89:103455.
- Achanccaray D, Hayashibe M (2020) Decoding Hand Motor Imagery Tasks Within the Same Limb From EEG Signals Using Deep Learn- ing. IEEE Transactions on Medical Robotics and Bionics 2: 692–699, 2020.
- Zhang R, Zong Q (2020) Hybrid Deep Neural Network Using Transfer Learning for EEG Motor Imagery Decoding. Elsevier Biomedical Signal Processing and Control 63:102144.
- Ozcan AR, Erturk S (2019) Seizure Prediction in Scalp EEG Using 3D Convolutional Neural Networks With an Image-Based Approach. IEEE Transactions on Neural Systems and Rehabilitation Engineering 27: 2284–2293.
- Ji J, Xing X, Yao Y, Li J, Zhang X. Convolutional Kernels With an Element-Wise Weighting Mechanism for Identifying Abnormal Brain Connectivity Patterns. Elsevier Pattern Recognition 109: 107570.
- Alazrai R, Abuhijleh M, Alwanni H, Daoud MI (2019) A Deep Learning Framework for Decoding Motor Imagery Tasks of the Sam Hand Using EEG Signals. IEEE Access 7: 612–627.
- Wang P, Jiang A, Liu X, Shang J. LSTM-Based EEG Classification in Motor Imagery Tasks. IEEE Transactions on Neural Systems and Rehabilitation Engineering 26: 2086–2095.
- Chen Z, Howe A, Blair H, Cong J (2018) CLINK: Compact LSTM Inference Kernel for Energy Efficient Neurofeedback Devices. International Symposium on Low Power Electronics and Design 1–6.
- Fares A, Zhong S.-h, Jiang J. (2020) Brain-Media: A Dual Conditioned and Lateralization Supported GAN (DCLS-GAN) Towards Visualization of Image-Evoked Brain Activities. ACM International Conference on Multimedia 1764–1772.
- Liang Y, Ma Y (2020) Calibrating EEG Features in Motor Imagery Classification Tasks with a Small Amount of Current Data Using Multisource Fusion Transfer Learning. Elsevier Biomedical Signal Processing and Control 62: 102101.
- Khalaf A, Akcakayam M ( 2020) A Probabilistic Approach for Calibration Time Reduction in Hybrid EEG-fTCD Brain-Computer Interfaces. Springer BioMedical Engineering OnLine 19:1–18.
- Haselsteiner E, Pfurtscheller G (2000) Using Time-Dependent Neural Networks for EEG Classification. IEEE Transactions on Rehabilitation Engineering 8: 457–463.
- Xiong X, Yu Z, Ma T, Wang H, Lu X et al., Classifying Action Intention Understanding EEG Signals Based on Weighted Brain Network Metric Features. Elsevier Biomedical Signal Processing and Control, 59:101893.
- Xiong X, Fu Y, Chen J, Liu L, Zhang X (2019) Single-Trial Recognition of Imagined Forces and Speeds of Hand Clenching Based on Brain Topography and Brain Network. Springer Brain topography 32:240–254.
- Jadav G, Lerga J, Sˇtajduhar I (2020) Adaptive Filtering and Analysis of EEG Signals in the Time-Frequency Domain Based on the Local Entropy. Springer Open Journal on Advances in Signal Processing 2020: 1–18.
- Rodrigues P LC, Jutten C, Congedo M (2018) Riemannian Procrustes Analysis: Transfer Learning for Brain-Computer Interfaces. IEEE Transactions on Biomedical Engineering 66: 2390– 2401.
- Wang Y, Qiu S, Ma X, He H. A Prototype-Based SPD Matrix Network for Domain Adaptation EEG Emotion Recognition. Elsevier Pattern Recognition 110:107626.
- Tzelepis C, Mezaris V, Patras I (2017) Linear Maximum Margin Classifier for Learning from Uncertain Data. IEEE Transactions on Pattern Analysis and Machine Intelligence 40: 2948– 2962.
- Georges N, Mhiri I, Rekik I (2020) Identifying the Best Data-Driven Feature Selection Method for Boosting Reproducibility in Classification Tasks. Elsevier Pattern Recognition 101:107183.
- Hekmatmanesh A, Wu H, Jamaloo F, Li M, Handroos H (2020) A Combination of CSP-Based Method With Soft Margin SVM Classifier and Generalized RBF Kernel for Imagery-Based Brain Computer Interface Applications. Springer Multimedia Tools and Applications1: 1-29.
- Dvorak D, Shang A, Abdel S, Suzuki W, Fenton A (2018) Cognitive Behavior Classification From Scalp EEG Signals. IEEE Transactions on Neural Systems and Rehabilitation Engineering 26: 729–739
- Song T, Zheng W, Song P, Cui Z (2018) EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks. IEEE Transactions on Affective Computing.
- Wang D, Yang J (2019) A Method of Expanding EEG Data Based on Transfer Learning Theory. International Conference on Networks, Communication and Computing 70–74.
- Zhang C, Wu D (2020) Applying Deep Learning for Decoding of EEG and BFV About Ischemic Stroke Patients and Visualization. International Conference on Machine Learning and Computing 89–95.
- Hosseini S, Guo X (2019) Deep Convolutional Neural Network for Automated Detection of Mind Wandering Using EEG Signals. ACM International Conference on Bioinformatics Computational Biology and Health Informatics 314–319.
- Xu G, Shen X, Chen S, Zong Y, Zhang C (2019) A Deep Transfer Convolutional Neural Network Framework for EEG Signal Classification. IEEE Access 7: 767– 776.
- Huang W, Xue Y, Hu L, Liuli H (2020) S-EEGNet: Electroencephalogram Signal Classification Based on a Separable Convolution Neural Network with Bilinear Interpolation. IEEE Access 8:636–646.
- Gao Z, Wang X, Yang Y (2019) EEG- Based Spatio-Temporal Convolutional Neural Network for Driver Fatigue Evaluation. IEEE Transactions on Neural Networks and Learning Systems 30: 2755–2763.
- Mandhana V, Taware R (2020) Classification of 3D Interpolated EEG Signals Using Hybrid R-3DCNN. ACM Symposium on Applied Computing 17–19.
- Xu M, Yao J (2020) Learning EEG Topographical Representation for Classification via Convolutional Neural Network. Elsevier Pattern Recognition 107390.
- S. Zhong, A. Fares, and J. Jiang, “An Attentional-LSTM for Improved Classification of Brain Activities Evoked by Images,” in ACM Inter- national Conference on Multimedia, 2019, pp. 1295–1303.
- Zhang G, Davoodnia V, Sepas-Moghaddam A (2019) Classification of Hand Movements From EEG Using a Deep Attention-Based LSTM Network. IEEE Sensors Journal 20: 3113–3122.
- Zheng X, Chen W, You Y, Jiang Y, Li M et al., Ensemble Deep Learning for Automated Visual Classification Using EEG Signals. Elsevier Pattern Recognition 102:107147.
- Zhang X, Yao L, Kanhere S, Liu Y, Gu T (2018) MindID: Person Identification From Brain Waves Through Attention-Based Recurrent Neural Network. ACM Conference on Interactive, Mobile, Wearable and Ubiquitous Technologies 2: 1–23.
- Lv Z, Qiao L, Wang Q, Piccialli F (2020) Advanced Machine-Learning Methods for Brain-Computer Interfacing. IEEE/ACM Transactions on Computational Biology and Bioinformatics
- Zhang Y, Nam C, Zhou G, Jin J, Wang X (2018) Temporally Constrained Sparse Group Spatial Patterns for Motor Imagery BCI. IEEE Transactions on Cybernetics 49: 3322–3332.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi