Review Article, J Appl Bioinforma Comput Biol Vol: 12 Issue: 6
Bioinformatics Advanced Tools to Study Antibiotic Resistant Bacteria and their Natural Products
Zahraa Ali Kamaz*
Department of Medical Science, University of Kerbala, Karbala, Iraq
- *Corresponding Author:
- Zahraa Ali Kamaz
Department of Medical Science,
University of Kerbala,
Karbala,
Iraq;
E-mail: biologyme1983@gmail.com
Received date: 04 May, 2023, Manuscript No. JABCB-23-97813;
Editor assigned date: 08 May, 2023, PreQC No. JABCB-23-97813 (PQ);
Reviewed date: 22 May, 2023, QC No. JABCB-23-97813;
Revised date: 04 July, 2023, Manuscript No. JABCB-23-97813 (R);
Published date: 11 July, 2023, DOI: 10.4172/2329-9533.1000290
Citation: Kamaz ZA (2023) Bioinformatics Advanced Tools to Study Antibiotic Resistant Bacteria and their Natural Products. J Appl Bioinforma Comput Biol 12:6.
Abstract
Antibiotics resistant bacteria are major health threats due to their rapidly growing number worldwide. Bacterial resistant genes are carried on plasmids, chromosomes, and transposons are transferred to non-resistant bacteria through horizontal genes transfer method and by integrons. Therefore, necessary strategies are required to control resistant bacteria spread, one strategy is developing new antibiotics as WHO announced, there is a need for 10 new drugs a decade to control resistant bacterial infection. However, the drug discovery approach is costly, and it requires long years of following research which slowed the process of drugs discovered in the last years. But the recent advances in gene sequencing technology have aided in searching for new drug candidates among bacteria and yet in the human microbiomes. In this review, a variety of bioinformatic tools are displayed in categories depending on their jobs to identify resistant genes and their target proteins, beside another set of tools to identify natural products among bacteria, furthermore, a new pipeline and workflow have been developed that will ease the research process for new drugs discovery.
Keywords: Antibiotics resistance; Natural products; Bioinformatic tools; Human microbiome; Resistant bacteria
Abbreviation:
AMR: Antimicrobial Resistant Bacteria; ARBs: Antibiotics Resistant Bacterias; ARGs: Mobile Resistant Genes; WHO: World Health Organization; FDA: US. Food and Drug Administration; ARCs: Antibiotics Resistant Cassettes; WGS: Whole Genome Sequences; EA Scores: Evolutionary Action Scores; GUI: Graphic based operating system Interface; DEG: Differential Genes Expression; KEGG: Kyoto Encyclopedia of Genes and Genomes; VFDB: Virulence Factor Database; HMM: Hidden Markov Model; NPs: Natural Products; BGCs: Biosynthetic Genes Clusters; PKS: Polyketide Synthase; NRPS: Non-Ribosomal Peptide Synthetase; RiPPs: Ribosomal synthesized and Post-translationally modified Peptides; NRPs: Non-Ribosomal Peptide synthetase; CDC: Center of Disease and Control prevention
Introduction
Bacteria resistance to antibiotics is a continuous problem that endangered the health of people around the world since bacteria’s ability to evolve and mutate rapidly to overcome harsh environmental burdens [1]. Therefore, the first resistant E. coli strain has been reported shortly after penicillin discovery and even before it’s been marketed in 1943, and the number of resistant bacteria is increasing substantially even new generations of antibiotics have been developed [2]. In a systematic survey for estimating Antimicrobial Resistance Bacteria (AMR) infection, mortality, and morbidity percentages, the survey covered 204 countries with 471 million individuals have been included, it’s been found that 1.27 million people are dying due to complications of AMR infections, another study reported about 35,000 deaths are attributed to AMR infections in US alone, and it’s been predicted to be about 10 million deaths by 2050 [3-5].
Bacteria can develop resistance by two main mechanisms, intrinsic factors which cause changes in bacteria membranes structures such as a mutation in LPS synthesis genes, activating drugs inhibitory enzymes, and drugs efflux through intracellular efflux pumps which are found in gram positive and gram negative bacteria, extrinsic mechanisms are those related to horizontal genes transfer between bacteria, Antibiotics Resistant Bacteria (ARBs) and mobile Resistant Genes (ARGs) transfer from farm animals to human, others factors are related to human intervention such as lacking of knowledge over prescribers and users, the absence of standard global controlling policies of antibiotics resistance spreads, and widespread prophylaxis use of antibiotics in food animals [6,7]. Bacteria are classified as resistant bacteria when it resists one agent in the antimicrobial classes, most common resistant pathogens that are reported by World Health Organization (WHO) in 2019 are E. coli, S. aureus, K. pneumoniae, S. pneumoniae, P. aeruginosa and A. baumannii, such widespread of AMR, put a further pressure for the need of developing new drugs, the pipeline for developing new drugs is a time-consuming and costly, it’s been estimated that invention a single antimicrobial agent requires at least 9 years of continuous work, furthermore, changes of US. Food and Drug Administration (FDA) guidelines for approving new drugs slowed the process of developing new antimicrobial agents and burdened pharmaceutical companies’ interest in drugs discovery research.
An urgent need for new drugs is necessary to combat AMR, therefore advanced technologies and tools in the drug discovery process are important to speed up the process and cut off the cost. Bioinformatic is a new multidisciplinary field that combines genomics, proteomics, and transcriptomics, bioinformatic has aided in the acceleration drugs discovery process through drug targets identification, estimation of drug biological features such as bioavailability, absorbance, toxicity, and blood-brain barrier permeability, and drugs side effects [8,9]. In this review, we will display the most recent bioinformatic tools to be used in resistant genes identification and screening, proteomics analysis, and new drugs discovery and drugs repurposing pipeline, that we believe such review will provide necessary information to resume drugs discovery research to tackle antibiotics resistant problem.
Literature Review
Bioinformatics tools advanced the discovery of resistant genes
Antibiotic-Resistant Genes (ARGs) are mobile elements can transfer between bacteria and from animals to humans, such rapid transfer mechanisms create a crisis for human health because of antibiotics resistance spread. AR genes can be found as free or in association with biological elements such as plasmids, integrons, and Antibiotics Resistant Cassettes (ARCs) which are integrated AR genes [10]. Huge amounts of biological data are now available in many repositories at free cost, which with the aid of bioinformatics tools enable researchers to screen resistant genes in the application for better treatment outcomes. Whole Genome Sequencing (WGS) data are applied in screening for resistome through real time surveillance, which has provided accurate data about Antibiotics Resistant Bacteria (ARBs) isolated from hospitals, and animals resources, comparing ARGs spread across countries, WGS is also been useful for epidemiological tracking of infectious diseases, define the genotypephenotype relationship, altogether such information is important in forming antibiotics controlling policies [11].
Bioinformatic tools for screening resistant genes associated with plasmids
For identification of small plasmids compromising of <10,000 contigs, blastn method is used to screen plasmids of interest against compass plasmid database, the database involves about 12,084 plasmids and the identification of resistant plasmids is based on replicon typing and MOB typing, usually the current database is useful for identification plasmids belongs mostly to proteobacteria and firmicutes [12]. For bacterial plasmids belonging to Enterobacteriaceae, blastn tool is used to align bacterial genome sequencing data against a specific plasmid database available at NCBI website, where plasmids in the query are identified based on identity and coverage percentage with a reference plasmid in the database, i.e., contigs with greatest identity >70% and coverage > 40% are identified as the same reference plasmid, contigs with lower coverage but it aligned to similar replicon type in the reference plasmids are collected together to create a draft plasmid which then analyzed through RAST server (Rapid Annotation of microbial genomes using Subsystems Technology) [13,14]. Another approach assigned to define ARGs associated with plasmids of Enterobacteriaceae bacteria, a combination of bioinformatics tools applied, plasmidfinder, a web tool that contains 559 plasmids types with full sequencing, the tool identifies plasmids of interest through searching WGS against the database, additional tool plasmidspades is used to screen plasmids that not identified by first one, the tool assembles short reads and contigs into a putative plasmid by calculating the copy number of plasmids and their coverage, thereafter, putative plasmids and plasmids are identified by plasmidfinder are blasted against reference plasmids available at NCBI database to identify their similar reference plasmids and finally a resfinder tool is applied to locate antimicrobial resistant genes [15-17]. For the detection of plasmids from diverse bacterial strains including environmental samples, a recent bioinformatic tool plasquid showed promising results regarding such purpose with high accuracy comparing to previously presented tools, the tool proved ability of detection plasmids from short read and circularized fragmented genomic materials [18].
Bioinformatic tools for the detection of ARGs associated with integrons
Integrons cassettes are mobile genetic materials flanked by a recombination site (attI) and an integrase gene (intI) whose sequences are vital for integrons classification into class 1,2,3,…besides it’s the main target for bioinformatics tools to identify integrons among resistant bacteria. Integron-resistant cassettes can be identified through a bioinformatic tool (in finder 0.1), a python integrated tool that asses the K-mer alignment score of aligned WGS with database sequences, the tool can identify class 1, 2, and 3 integrons, a second tool called the Integron Visualization and Identification Pipeline (I-VIP v1.2) is used to screen and annotates class 1 integrons [19,20]. An updated version of intergron finder 0.2 has been launched in 2022, the tool is built in python 3.7, and it’s more efficient than the previous version of detecting class I integrons, the tool works by searching for homologs sequences of integron class 1 in integrated datasets containing 21,105 complete genomes that allocated from NCBI websites, then after, ARGs are annotated by searching four databases are card, arg-annot, AMRfinderplus, and resfinder. For robustness results, another platform called integrall 1.2 is searching for homology sequences of intI among uncultured and cultured bacteria, it has a depository of more than 4800 nucleotides sequences of different bacteria, it provides quick results by blasting the provided FASTA sequences with additional features such as integrons nomenclature, genes cassette’s location and functionality.
Bioinformatic tools for resistome surveillance
Resistome term is applied for the collection of Antibiotic Resistant Genes (ARGs) and protoresistant genes that are isolated from bacteria communities including pathogenic and non-pathogenic bacteria. Antibiotics resistome is a dynamically expanding community of bacteria, and its contribution to diseases complication, has placed a global burden to detect their presence in different environments including the human body, an array of bioinformatics tools is developed for deep ARGs searching and defining their resources ARGs searching platform (ARGs-OSP) is comprising of more than fifty five thousands bacteria genomes sequences (complete and partial), and metagenomics datasets, ARGs-OSP pipeline can detect ARGs on bacterial genomes, plasmids and integrins.
Other bioinformatics tools are working on short read sequences and very long read sequences of metagenomics data such as DeepARG-SS and NanoARG, additional tools can be found in the same resource, however, all mentioned tools are error prone methods since it depends on the best hit, therefore, applying more than one tool for ARGs detection is more convenient to exclude false negative results. At the current time, there are no tools for the detection of pro-resistant genes (genes that show their resistance upon changing environmental conditions or it’s also known as silent resistant genes), a new method has been developed to detect such genes, the method is depending on measuring evolutionary action scores (EA scores) for genes of interest, EA scores are rationally correlated with the functional impact of codons variation of a gene, mutated genes with a functional impact usually have high EA score.
Bioinformatics for proteomics studies
Proteins are essential for cellular functions and growth and constitute most drugs targets in the drugs discovery pipeline, therefore, applying omics tools have accelerating the drugs discovery process, also it reduces the cost of drugs discovery research by data mining through taking advantage of in silico studies before clinical trials. Machine learning algorithms have expanded small molecules searching by identifying best interacting molecules with proteins targets, identifying proteins functions and structures, binding domains, protein-protein interactions, are all become feasible pipelines with the aid of bioinformatic tools nevertheless, proteins are not single molecules, rather than are complexes in biological functions, to explore interactome in pathogens and non-pathogens, numerous tools are developed to fill the gap of proteomics field, now days, tools for proteins network building and analysis are free to use, furthermore proteins databases which involve thousands of known proteins with homolog blasting technology that aid in leveraging information about de novo proteins as new targets, all above technologies are important to break down the cycle of identifying new target proteins which was in previous times rely on known proteins only.
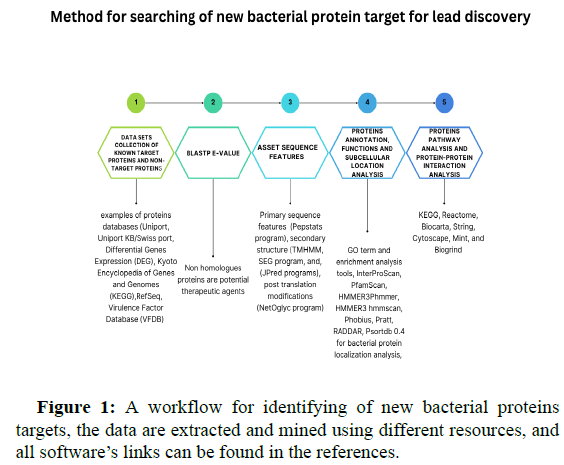
Bioinformatics tools for identifying new bacterial drugs target proteins
It has been known from the time of antibiotics invention, bacterial membranes and subcellular proteins are the main targets where antibiotics interact and confer their actions, at the same time, and bacteria are evolving survival mechanisms by changing the structure of target proteins. Exploring new target proteins is essential to overcoming bacterial resistance, set of tools are designed to for search and identify new target proteins. A new pipeline Figure 1 is constructed based on the literatures review to identify new target proteins, it’s occupied with essential tools required for each step of proteomics analysis. Since proteins are not single entity molecules but function in complexes, it’s necessary to display some of the tools that apply machine learning to identify core proteins and virulence proteins, net confer is a Graphic based operating system interface (GUI) based tool which used for comparative proteins’ networks analysis, the tool analysis is based on comparing several parameters such as find common nodes and edges, identify key nodes by calculation of degrees, between’s centrality, hub and authority scores and other measurements.
Bioinformatics for identifying bacterial virulence factors
Virulence factors are important pathogenic markers that bacteria develop to cause diseases, virulence factors are proteins located on either bacterial membrane or secretory intracellular factors, which are categorized according to the Virulence Factors Database (VFDB) into adherence proteins, invasion proteins, secretory systems, flagella, exotoxins, exoenzymes, immune modulators, biofilm, metabolic regulators, post-translation regulators, stress response proteins, antimicrobial agents, and others. Searching for and identifying virulence factors in bacteria is important for developing therapeutic agents and vaccines, bioinformatic field has tremendously accelerated drugs discovery research, with the advent of the availability of virulence factor database, blast searching for orthologues virulence factor sequences, motif sequences, signal sequences, and domain sequences among different bacteria are becoming straightforward task, however, blast search results are not accurate mostly and the need of more accurate tools are essential. From the literatures reviewing, patho fact software is a python based pipeline for prediction of virulence factors, the prediction is based on two steps, the first step is searching for homologs query sequences against Hidden Markov Model (HMM) profiles which include datasets retrieved from VFDB website, and the second step is implanting random forest model for creating classifier trees.
Bioinformatic tools for drug discovery in bacteria
Secondary metabolites and natural products with antimicrobial activities have encompassed much interest in the drug discovery field, since penicillin has been isolated from bacteria and till now, biotechnological approaches for drug discovery have been changed tremendously, previously culture methods were applied for searching of microbes’ biomolecules accompanied with ethanol extraction, compound isolation, and purification. However, this method is costineffective and impractical since it requires optimization of the culture environment for every single microbe. Recent advances in high throughput genome sequences made the roadmap of Natural Products (NPs) discovery from genes to molecules amenable, easy to apply, and cost-effective task. It’s noteworthy that mentioning the availability of many bioinformatic tools and databases for NPs discovery from bacteria and fungi, furthermore, other algorithms are built to identify novel NPs with efficient expression without rediscovery of known compounds or compounds with low quantities. More recently, genome mining technology has revolutionized the process of NPs discovery the term is applied for steps of identifying genes involved in bioactive molecules biosynthesis, isolation, and purification, for which omics tools are synchronized to ease aforementioned process.
Discussion
Genome mining and bioinformatics
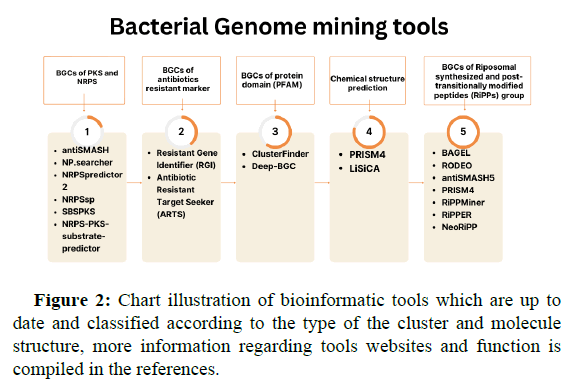
While the complete genome sequence of Streptomces coelicolor has been published in 2002, more silent secondary products are discovered by applying a genome mining strategy. The process which is merely dependent on identifying Biosynthetic Gene Clusters (BGCs) that encode producing molecules through microbial genome or community, although the huge variability of BGCs among microbes, there are signature enzymes (core enzymes) that are conserved and the best examples are Polyketide Synthase (PKS) and Non-Ribosomal Peptide Synthetase (NRPS) as well as tailor enzymes for targeting cryptic biosynthetic pathways. Several mining methods are devoted by scientists to help in minimizing hit and find fail since genome mining tools are largely trained on sequence similarity search, one method is mining bioactive features of target compounds through searching for bioactive moiety in BGCs, examples are numerous including applying bioactive chemical features, and ligand binding feature, once the bioactive molecule is identified, another mining type is through searching for analog compounds, and one another strategy is antibiotic resistant genes mining, microbes which produce active antibiotics are usually develop a duplicate copy of resistant genes to avoid selftoxication, such resistant genes are the hook for mining tools. The next step following BGCs identification is linking it to the desired metabolites, two approaches are now available to help in finding the metabolite pathway, one is the utilization of mass spectrometry for the isolation of producing compounds from crude fermentation extracts of microbes, on the other hand, the second approach is for non-cultivable microbes which depend on metabolite structure prediction and synthesis. Bioinformatic tools for genome mining and metabolite structure prediction are categorized into groups according to their function and are illustrated in Figure 2.
Bioinformatics approach for mining human microbiome
Human microbiome is comprised of a large community of bacteria that harbored bioactive genes with multiple functions, cross-talk of gut bacteria to bacteria and to their host has established the fact of their role in human health since the century of human microbiome discovery, human microbiome provides our bodies with vitamins and fatty acids, also it has been confirmed their sources of novel secondary metabolites. Because most microbiome bacteria are tedious in their cultural requirements, genomic and metagenomic sequencing technology have advanced microbiome lead discovery and the knowledge of their responsible bioactive genes. However, other efforts have fostered techniques for culturing recalcitrant bacteria such as using natural enlivenment simulation approach. There are several branches have been developed for applying metagenomic sequencing technology for microbiome mining, one is based on searching DNA sequencing libraries of human microbiome databases (human microbiome NIH project, microbiome DB, GMrepo V2, and others) and predicting BGCs that produce bioactive compounds, such approach have led to discover novel compounds such as lactocillin with selective bactericidal activity against gram positive bacteria without affecting vaginal beneficial microbiome bacteria, humimycins with antibacterial activity have discovered by searching for known (non-ribosomal peptides) NRPs in BGCs pool of gut bacteria then synthesized the identified NRP in the lab, another novel compounds dipeptide aldehydes with anti-cathepsins proteases activity were discovered through identifying a unique distribution pattern of BGCs in human gut bacteria. The second approach of microbiome mining is functional metagenomic which rely on construction of cosmid DNA libraries mostly in E. coli utilizing human microbiome metagenomic databases, then searching for bioactive effectors through homologues sequence alignment method, bioinformatic tools for searching of bioactive effectors are many, and are either dependent on sequence similarity comparison of known effectors or distribution probability of effector proteins by enrichment machine learning method with set of positive and negative effectors samples, some examples of bioinformatic tools are sieve which predicts effectors of T3SS system, T3MM which applied markov model for prediction, bean 0.2 for prediction of T3SS effectors through sequence similarity approach, S4TE for prediction of T4SS effectors proteins, and effective DB for prediction effectors from multiple bacterial secretion systems. Another approach for microbiome mining is the chemical analysis of metabolites from human bacteria by mass-spectrometry technology MS, the method by which known reference samples are provided to identify microbiome metabolites.
Overview of recent updates of antibiotic resistant bacteria
To address the need for more tools to fight bacterial resistance, understanding their risks, evolving rates, and death rates are essential to improve health wellness and the economy, most recent report of MR bacteria is the Center of Disease and Control prevention (CDC) report in 2019 that estimated about 2.8 million infections were reported due to antibiotic resistant bacteria each year, besides about 35,000 deaths, and top notch resistant bacteria were drug resistant Neisseria gonorrhea with 550,000 infections. While another survey study which conducted to estimate death cause of AMR in 204 countries, estimated death rate in 2019 was 4.95 million at 95% UI, the highest death rate was in sub-Saharan African countries and the lowest in Australia, top notch resistant pathogens are those attributed with lower respiratory tract infection with 1.5 million infections in 2019. In European countries, a report of antibiotic resistance conducted from 2010-2013 recorded multi drug resistant Klebseilla pneumoniae percentage has increased significantly, on the other hand, methicillin resistant Staphylococcus aureus infections have decreased significantly, in the same report, most European regions that harbor AMR infections were southern and Southern-Eastern parts recent updates on antibiotic resistance burden at global level are missing though WHO announcement of AMR global death to be more than 10 million in 2050. This huge number of resistant bacteria infections and their economic impacts globally place a prioritization need for unified global strategies to fight antibiotics resistance including developing new drugs.
Conclusion
Although antibiotic resistant bacteria are developing rapidly each year, overall designated budget for antibiotic resistant research is low, whereas, WHO is assumed about 10 new drugs a decade are required to tackle antibiotic resistance globally with an estimated budget of 2 million USD each year. The time scale for developing a new drug which is about 10 years is the most common challenge in the drug research field, however, the advances in genomic and metagenomic fields have reduced the time scale of drug developing into 2 years.
Nowadays, many bioinformatic algorithms and databases are available to search for bacterial resistant genes and their proteomes, moreover, the detection of cryptic resistant genes and target proteins is becoming feasible. Also, there are bioinformatic tools for searching of secondary metabolites with antibacterial activity from a variety of sources including large bacterial communities such as human microbiome. Despite such many bioinformatic tools, the tools for identifying de novo bacterial biosynthetic genes and their related pathways are still missing.
Ethical Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Materials
Not applicable.
Competing Interest
I am as the author declares that I have no competing interest in the current work.
Funding
I am confirming this current work is not received any funding from any institution including that I work in.
Author’s Contribution
I am the only author for the current work who responsible for conceptualization, ideas, study design, and writing.
Acknowledgments
I am here acknowledging AL-Zahraa university president and staff for their collaboration to keep the house environment for the current work.
References
- Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, et al. (2022) Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399:629-655.
[Crossref] [Google Scholar] [PubMed]
- Imchen M, Moopantakath J, Kumavath R, Barh D, Tiwari S, et al. (2020) Current trends in experimental and computational approaches to combat antimicrobial resistance. Front Genet 11:563975.
[Crossref] [Google Scholar] [PubMed]
- Gogry FA, Siddiqui MT, Sultan I, Haq QMR (2021) Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front Med (Lausanne) 8:677720.
[Crossref] [Google Scholar] [PubMed]
- Xu C, Kong L, Gao H, Cheng X, Wang X (2022) A review of current bacterial resistance to antibiotics in food animals. Front Microbiol 13:822689.
[Crossref] [Google Scholar] [PubMed]
- Xia X (2017) Bioinformatics and drug discovery. Curr Top Med Chem 17:1709-1726.
[Crossref] [Google Scholar] [PubMed]
- Ramharack P, Soliman MES (2018) Bioinformatics based tools in drug discovery: The cartography from single gene to integrative biological networks. Drug Discov Today 23:1658-1665.
[Crossref] [Google Scholar] [PubMed]
- McMillan EA, Gupta SK, Williams LE, Jove T, Hiott LM, et al. (2019) Antimicrobial resistance genes, cassettes, and plasmids present in Salmonella enterica associated with United States food animals. Front Microbiol 10:832.
[Crossref] [Google Scholar] [PubMed]
- Mellmann A, Bletz S, Boking T, Kipp F, Becker K, et al. (2016) Real time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol 54:2874-2881.
[Crossref] [Google Scholar] [PubMed]
- Douarre PE, Mallet L, Radomski N, Felten A, Mistou MY (2020) Analysis of compass, a new comprehensive plasmid database revealed prevalence of multireplicon and extensive diversity of IncF plasmids. Front Microbiol 11:483.
[Crossref] [Google Scholar] [PubMed]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, et al. (2014) The seed and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42.
[Crossref] [Google Scholar] [PubMed]
- Kudirkiene E, Andoh LA, Ahmed S, Herrero-Fresno A, Dalsgaard A, et al. (2018) The use of a combined bioinformatics approach to locate antibiotic resistance genes on plasmids from whole genome sequences of Salmonella enterica serovars from humans in Ghana. Front Microbiol 9:1010.
[Crossref] [Google Scholar] [PubMed]
- Carattoli A, Zankari E, Garcia-Fernandez A, Larsen MV, Lund O, et al. (2014) In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895-3903.
[Crossref] [Google Scholar] [PubMed]
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, et al. PlasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics 2016;32:3380-3387.
[Crossref] [Google Scholar] [PubMed]
- Deng Y, Bao X, Ji L, Chen L, Liu J, et al. (2015) Resistance integrons: Class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob 14:38-45.
[Crossref] [Google Scholar] [PubMed]
- Torres-Elizalde L, Ortega-Paredes D, Loaiza K, Fernandez-Moreira E, Larrea-Alvarez M (2021) In silico detection of antimicrobial resistance integrons in Salmonella enterica isolates from countries of the andean community. Antibiotics 10:1388.
[Crossref] [Google Scholar] [PubMed]
- Neron B, Littner E, Haudiquet M, Perrin A, Cury J, et al. (2022) Integron finder 2.0: Identification and analysis of integrons across bacteria, with a focus on antibiotic resistance in Klebsiella. Microorganisms 10:700.
[Crossref] [Google Scholar] [PubMed]
- Moura A, Soares M, Pereira C, Leitao N, Henriques I, et al. (2009) Integrall: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096-1098.
[Crossref] [Google Scholar] [PubMed]
- Kim DW, Cha CJ (2021) Antibiotic resistome from the one health perspective: Understanding and controlling antimicrobial resistance transmission. Exp Mol Med 2021 53:301-309.
[Crossref] [Google Scholar] [PubMed]
- Peng Z, Mao Y, Zhang N, Zhang L, Wang Z, et al. (2021) Utilizing metagenomic data and bioinformatic tools for elucidating antibiotic resistance genes in environment. Front Environ Sci 9:507.
- Marciano DC, Wang C, Hsu T-K, Bourquard T, Atri B, et al. (2022) Evolutionary action of mutations reveals antimicrobial resistance genes in Escherichia coliNat Commun 13:1-13.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi