Research Article, J Appl Bioinforma Comput Biol Vol: 9 Issue: 5
Computer-Aided Probing of the Pathogenic SNPs of Spartin Protein Associated with Hereditary Spastic Paraplegia
Ammara Akhtar*, Sobia Nazir Ch and Mureed Hussain
Department of Life Sciences, University of Management and Technology, Lahore, Pakistan
*Corresponding Author: Ammara Akhtar
Department of Life Sciences, University of Management and Technology, Lahore, Pakistan, 54770
E-mail: ammaraakhtar3@gmail.com
Received: Aug 08, 2020 Accepted: Sep 26, 2020 Published: Oct 04,2020
Citation: Akhtar A, Ch SN, Hussain M (2020) Computer-Aided Probing of the Pathogenic Snps of Spartin Protein Associated with Hereditary Spastic Paraplegia . J Appl Bioinforma Comput Biol 9:5. doi: 10.37532/jabcb.2020.9(5).181
Abstract
Background: Hereditary Spastic Paraplegia (HSP) is a neurological disorder associated with causing progressive spasticity in the lower limb of humans. In this study, the computational analysis was limb of humans. In this study, the computational analysis was performed to screen out the pathogenic missense and splicing variants of Spartin. The mutations in this mitochondrial protein can subsequently lead to HSP. Method: To discover novel mutations of Spartin protein, the missense and variants were obtained from gnomAD (Genome Aggregation Database), and further subjected to CADD (Combined Annotation Dependent Depletion) analysis. To validate the results, it was compared with various in silico mutation analysis tools. To accomplish novelty in the recent work, the mutations were analyzed and compared with the web- based tool ClinVar for finding novel mutations. Results: After stringent analysis, 3 missense mutations and 4 spicing mutations were obtained which have not been previously reported in any kind of databases or scientific work, thus can be considered as novel.
Keywords: Hereditary spastic paraplegia; nsSNPs; In silico; Variant, Spartin; gnomAD; CADD; Variation
Introduction
HSP (also known as Strumpell-Lorrain disease) is neurodegenerative disorder associated with the axonal-degeneration [1]. Some proteins involved in HSP interfere with more than one normal processes like axonal movement, metabolism of lipids, macroautophagy, mitochondrial functioning, myelination, endoplasmic reticulum, corticospinal tract, protein folding and processing of microtubules [2]. The prevalence rate of hereditary spastic paraplegia is highly variable globally. The global prevalence of the hereditary spastic paraplegia is estimated to be 4.26/100,000, mainly depending upon the route of inheritance as well as the specific geographic regions being affected [3].
The Spartin is a multifunctional protein which is naturally localized in the mitochondria [4]. Troyer syndrome is predominantly an Autosomal Recessive (AR) type of hereditary spastic paraplegia, which is caused by the mutations in the Spartin protein. It is basically a disorder primarily associated with various complications and clinical features like dysarthria, short heighted, distal amyotrophy, delay in the normal development and additionally spasticity. The syndrome is caused by pre-mature truncation of protein, predominantly due to known frameshift mutation, “1110delA” on exon number 4, at chromosome position 13q. This mutation lead to the formation of dysfunctional protein, unable to perform its normal function [5].
Identifying novel SNPs associated with a disease pose major challenges in the genomic analysis [6]. Each individual chiefly comprises of a vast variety of variants for a specific protein, according to an estimate, mainly 12,000-14,000 [7]. The selection of pathogenic variants among a list of thousand variants is a major challenge in the modern genetic analysis [8]. With the advancement of digital technology, the insilico approaches are significantly acclaimed for the prediction and investigation of different biological processes and pathways [9].
This study was carried out to identify novel SNPs of SPG20 gene encoding for spartin protein with the help of bioinformatic tools.
Materials and Methods
Variant Retrieval
The list of variants was retrieved through online genetic databases, gnomAD and Variation View. The protein sequence of spartin was taken from NCBI (National Center for Biotechnology Information).
Variants Selection
Different filters were applied to obtain missense and splicing variants for further analysis. An allelic frequency filter list of <0.002 was applied on the variant list. After filtering, the remaining variants were subjected to CADD Analysis, which provides C-score value of each variant. C- score provide information about highly pathogenic, likely or moderately pathogenic and low pathogenic variants [10]. A C-score filter was applied by setting a cut off value of ≥15. After filtering, remaining variants were compared with various in silico tools to validate our results.
Missense analysis
For missense analysis, the selected variants were further subjected to variant analysis against a range of bioinformatic tools PHD- SNPg (≥ 0.5 = Pathogenic) : SNP&GO (>0.5 = disease): PROVEAN (>-2.5 = neutral, <-2.5= pathogenic): UMD-PREDICTOR (<50 polymorphism; (ii) 50–64 probable polymorphism; (iii) 65–74 probably pathogenic mutation and (iv) >74 pathogenic mutation): PredictSNP2. The missense variants were checked in the ClinVar.
Stability analysis
For stability analysis, a cut off value was applied for each tool (PHD-SNPg ≥0.9: PROVEAN ≤-4.00: Predict-SNP2 ≥0.8: SNP&GOs ≥0.6: UMD-Predictor ≥80), those variants fulfilling the given criteria were analyzed in I-Stable.
Splicing Variant analysis
Variants having the potential to break or alter the splicing site associated with spartin protein were analyzed by utilizing different tools, “Human Splice Finder (HSF), Spliceman and SPICE”. To find novel mutations in this study, the mutations were checked in the ClinVar.
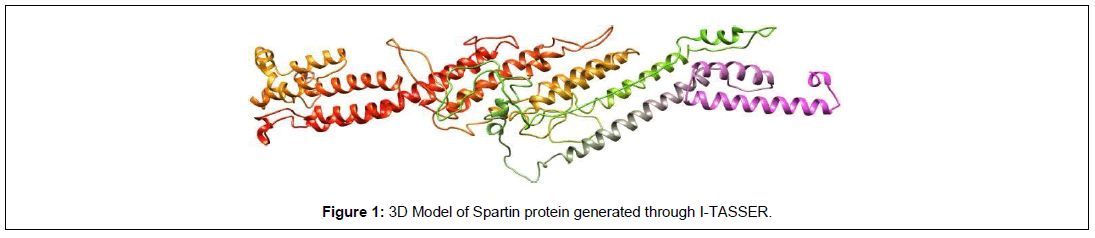
3D Modelling of Spartin
The 3D modelling of protein was performed to analyze the effect of mutations on the structure and function of the protein. The model of protein was built through I-TASSER. The best model was selected showing high confidence level, depending upon the values of RMSD and C-scoring [11].
Mutation effect on protein stability
The mutations of the spartin protein were visualized within Chimera by inducing changes in the target sequence of protein. The Chimera tool do not provide any significant prediction related to the effect of the mutation on the structure and stability of the protein. So, for this purpose, in silico tool, Mutpred2 were used.
Conservationanalysis ofpathogenic nsSNPs inSpartin
ConSurf was used to calculate the conservation score of the amino acids of spartin protein. ConSurf combines the conservation information of protein with its structural and functional properties, and finally predict the conservation status [12].
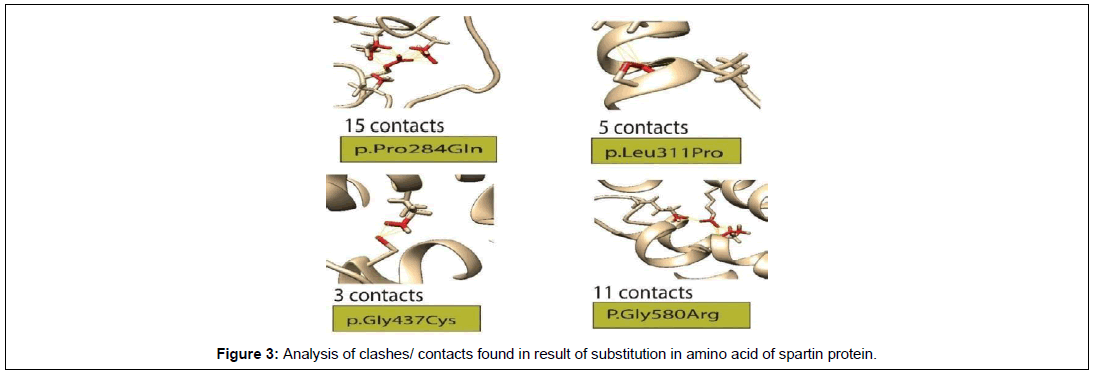
Clashes and contacts
Clashes/contacts associated with the amino acid residue of the protein were studied on the basis of VDW radii. Clashes/contacts provide significant information about the effect of mutations leading to unfavorable interaction within the protein structure [13].
Hydrophobicity analysis
Chimera was used to study the hydrophobicity properties of the amino acid residues based on kdHydrophobicity scale.
Result
3D modelling of Spartin
The structural models of proteins were obtained through I- TASSER. In the recent study, among five different models generated through I-TASSER, the first one was selected as the best model of spartin protein (Figure 1). For SPG20, the selected model has C- score=-1.03, estimated TM-score = 0.58±0.14, estimated RMSD = 10.4±4.6Å [11].
Missense variant analysis of SPG20
Variants of SPG20 were obtained through gnomAD. Total 646 variants were retrieved after applying filters. A filter of allelic frequency of < 0.002 was further applied and remaining 643 variants were analyzed in the CADD. In CADD results, filter of missense and C score ≥ 15 were used. After applying filters, 294 variants were achieved.
Pathogenic missense nSNPs analysis through various in silico tools
The remaining 294 variants were subjected to various web-based tools for mutation analysis like SNPs&GO, PHD-SNPg, PREDICT- SNP2, PROVEAN and UMD-Predictor. The distinct set of filters “deleterious or pathogenic or probably pathogenic” were used. A cut off value was applied for every tool (PHD.SNPg (0.9): PROVEAN (-4.00): SNP&GO (0.6): Predict SNP2 (0.8): UMD-predictor (80), 9 variants were procured (Table 1).
| Chr | Pos | R | A | Substitution | PhD-SNPg | PROVEAN | PredictSNP2 | SNP&GO | UMD | Phred | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre | S | Pre | S | Pre | S | Pre | S | Pre | S | ||||||
| 13 | 36878716 | T | C | p.Asn596Ser | P | 0.969 | D | -4.1 | D | 1 | Dis | 0.784 | 81 | P | 29.3 |
| 13 | 36909408 | T | C | p.Tyr187Cys* | P | 0.988 | D | -4.1 | D | 1 | Dis | 0.532 | 93 | P | 20.4 |
| 13 | 36909178 | G | A | p.Arg264Cys | P | 0.933 | D | -4.4 | D | 1 | Dis | 0.528 | 96 | P | 22.8 |
| 13 | 36909900 | G | T | p.Ala23Asp* | P | 0.985 | D | -4.5 | D | 1 | Dis | 0.628 | 93 | P | 15.8 |
| 13 | 36905729 | C | T | p.Cys272Tyr | P | 0.99 | D | -4.5 | D | 1 | Dis | 0.794 | 90 | P | 22.9 |
| 13 | 36878620 | T | C | p.Tyr628Cys | P | 0.963 | D | -4.6 | D | 1 | Dis | 0.599 | 99 | P | 31 |
| 13 | 36886488 | C | T | p.Gly537Asp | P | 0.986 | D | -4.8 | D | 1 | Dis | 0.613 | 90 | P | 27.9 |
| 13 | 36909259 | C | A | p.Gly237Trp | P | 0.993 | D | -5.3 | D | 1 | Dis | 0.698 | 96 | P | 22.3 |
| 13 | 36905612 | A | G | p.Leu311Pro | P | 0.993 | D | -5.3 | D | 1 | Dis | 0.667 | 84 | P | 23.4 |
| 13 | 36909282 | T | A | p.Gln229Leu | P | 0.991 | D | -5.4 | D | 1 | Dis | 0.571 | 93 | P | 22.2 |
| 13 | 36888546 | A | C | p.Val434Gly | P | 0.989 | D | -5.6 | D | 1 | Dis | 0.509 | 90 | P | 25.3 |
| 13 | 36905693 | G | T | p.Pro284Gln | P | 0.993 | D | -5.9 | D | 1 | Dis | 0.552 | 84 | P | 23 |
| 13 | 36886360 | A | C | p.Val552Gly | P | 0.974 | D | -6.2 | D | 1 | Dis | 0.59 | 90 | P | 28.5 |
| 13 | 36905621 | C | T | p.Gly308Glu | P | 0.982 | D | -6.4 | D | 1 | Dis | 0.54 | 87 | P | 23.3 |
| 13 | 36888525 | C | T | p.Gly441Asp | P | 0.989 | D | -6.4 | D | 1 | Dis | 0.809 | 90 | P | 25.7 |
| 13 | 36878765 | C | T | p.Gly580Arg | P | 0.979 | D | -6.4 | D | 1 | Dis | 0.852 | 81 | P | 28.8 |
| 13 | 36909294 | G | A | p.Pro225Leu | P | 0.905 | D | -6.5 | D | 1 | Dis | 0.599 | 84 | P | 22 |
| 13 | 36905666 | T | C | p.Tyr293Cys | P | 0.913 | D | -6.6 | D | 1 | Dis | 0.515 | 93 | P | 23.1 |
| 13 | 36888538 | C | A | p.Gly437Cys* | P | 0.95 | D | -7.7 | D | 1 | Dis | 0.661 | 100 | P | 25.3 |
Table 1: List of high-risks variants of Spartin (Abbreviations: D: Deleterious, Dis: disease, P: pathogenic, Pre: Prediction, S: Score).
Stability Predictions
The effect of mutations on the stability, structure and function of the spartin protein was observed through I-Stable. A “decrease” filter was applied in the result obtained after stability analysis. Total 9 variants were obtained after analysis, which has been visually illustrated in the Figure 2 and Table 2.
| Chr | Pos | Ref | Alt | Substitution | i-Mutant | DDG | Mupro | Conf. Score | iStable | Conf. Score |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 36878716 | T | C | p.Asn596Ser | Decrease | -0.11 | Decrease | -1 | Decrease | 0.861227 |
| 13 | 36909408 | T | C | p.Tyr187Cys* | Decrease | -1.2 | Decrease | -0.2389 | Decrease | 0.749386 |
| 13 | 36909178 | G | A | p.Arg264Cys | Decrease | -0.95 | Decrease | -1 | Decrease | 0.758092 |
| 13 | 36909900 | G | T | p.Ala23Asp* | Decrease | -0.95 | Decrease | -1 | Decrease | 0.758092 |
| 13 | 36905729 | C | T | p.Cys272Tyr | Decrease | -0.17 | Decrease | -0.5574 | Decrease | 0.838062 |
| 13 | 36905612 | A | G | p.Leu311Pro | Decrease | -1.81 | Decrease | -1 | Decrease | 0.863248 |
| 13 | 36905693 | G | T | p.Pro284Gln | Decrease | -1.91 | Decrease | -0.68 | Decrease | 0.759634 |
| 13 | 36878765 | C | T | P.Gly580Arg | Decrease | -0.23 | Decrease | -0.614 | Decrease | 0.82841 |
| 13 | 36888538 | C | A | p.Gly437Cys* | Decrease | -1.55 | Decrease | -0.0557 | Decrease | 0.769476 |
Table 2: List of high-risks variants of Spartin (Abbreviations: D: Deleterious, Dis: disease, P: pathogenic, Pre: Prediction, S: Score).
MutPred Predictions of mutation effect on functional and structural properties of spartin
The 9 nSNPs were analyzed further in the web-based server Mutpred to study the effect of AAS on the structure and function of the spartin protein. The predictions exhibited that these SNPs have the potential to disrupt the structural and functional properties of normal spartin protein (Table 3).
| Chr | Pos | Substitution | Effect |
|---|---|---|---|
| 13 | 36878716 | p.Asn596Ser | Altered Ordered interface (0.29) |
| Gain of Relative solvent accessibility (0.29) | |||
| Altered Metal binding (0.28) | |||
| Gain of Strand (0.27) | |||
| Altered Transmembrane protein (0.23) | |||
| Loss of Allosteric site at N596 (0.23) | |||
| Loss of Catalytic site at N596 (0.22) | |||
| Altered DNA binding (0.20) | |||
| Gain of O-linked glycosylation at S593 (0.12) | |||
| Loss of GPI-anchor amidation at N596 (0.02) | |||
| 13 | 36909408 | p.Tyr187Cys | Loss of Phosphorylation (0.36) |
| Gain of Loop (0.28) | |||
| Loss of Strand (0.26) | |||
| 13 | 36909178 | p.Arg264Cys | - |
| 13 | 36909900 | p.Ala23Asp | Altered Disordered interface (0.28) |
| Gain of Relative solvent accessibility (0.25) | |||
| Gain of Acetylation at K22 (0.25) | |||
| 13 | 36905729 | p.Cys272Tyr | Gain of Loop (0.27) |
| Gain of Sulfation at C272 (0.02) | |||
| 13 | 36905612 | p.Leu311Pro | Gain of Intrinsic disorder (0.32) |
| Altered Transmembrane protein (0.14) | |||
| 13 | 36905693 | p.Pro284Gln | Loss of ADP-ribosylation at R282 (0.22) |
| Altered Transmembrane protein (0.10) | |||
| 13 | 36878765 | P.Gly580Arg | Altered Ordered interface (0.38) |
| Altered Transmembrane protein (0.31) | |||
| Altered Disordered interface (0.30) | |||
| Gain of Relative solvent accessibility (0.30) | |||
| Altered Metal binding (0.27) | |||
| 13 | 36888538 | p.Gly437Cys | Altered Transmembrane protein (0.24) |
| Altered Disordered interface (0.18) |
Table 3: MutPred predictions.
Amino acids conservation profile by in silico tool ConSurf
The conservation analysis of 9 variants was carried out through ConSurf. The result exhibited that amino acid residues Asn596, Tyr187, Arg264, Pro284, Gly580 were functional residues so exposed, while Cys272, Ala23, Gly437, Leu311 were structural residues, it means they will be probably buried in the protein structure. The results predicted that these 9 nsSnps have the potential to damage the structure and function of the spartin protein (Table 4).
| Chr | Pos | Ref | Alt | AAS | Conservation Score | Prediction |
|---|---|---|---|---|---|---|
| 13 | 36878716 | T | C | p.Asn596Ser | 9 | Highly conserved and exposed (f) |
| 13 | 36909408 | T | C | p.Tyr187Cys* | 8 | Highly conserved and exposed (f) |
| 13 | 36909178 | G | A | p.Arg264 | 6 | Highly conserved and exposed (f) |
| 13 | 36909900 | G | T | p.Ala23Asp* | 9 | Highly conserved and buried (s) |
| 13 | 36905729 | C | T | p.Cys272Tyr | 7 | Highly conserved and buried (s) |
| 13 | 36905612 | A | G | p.Leu311Pro | 8 | Highly conserved and buried (s) |
| 13 | 36905693 | G | T | p.Pro284Gln | 9 | Highly conserved and exposed (f) |
| 13 | 36878765 | C | T | P.Gly580Arg | 8 | Highly conserved and exposed (f) |
| 13 | 36888538 | C | A | p.Gly437Cys* | 8 | Highly conserved and buried (s) |
Table 4: Consurf results predicting the conservation of the amino acids in the Spartin protein.
Contact and clashes analysis
For Clashes and contacts analysis, the protein structure with amino acid substitution were analyzed through Chimera. Out of 9, total 4 missense mutations showed diverse kind of interactions with the nearest residues. The contacts and clashes have the potential to disrupt the normal structure and function of the spartin protein (Figure 3).
Hydrophobicityanalysisofspartinaminoacidresidues
Hydrophobicity analysis was performed by using Chimera. During analysis, amino acid residues were represented with several characteristical attributes based on kdHydrophobicity. Each amino acid was assigned a particular value according to a scale provided by Kyte and Doolittle. A color was allotted to protein residues based on their hydrophilic or hydrophobic properties (Blue color exhibit (most hydrophilic): White color (Neutral) and orange red (most hydrophobic) as well as a value based on the hydropathy scale (more positive value means hydrophobic and more negative means amino acid have hydrophilic properties) (Table 5 and Figure 4).
| Chr | Pos | Ref | Alt | AAS | Hydrophobicity value | |
|---|---|---|---|---|---|---|
| Wild | Mutant | |||||
| 13 | 36878716 | T | C | p.Asn596Ser | -3.5 | -0.8 |
| 13 | 36909408 | T | C | p.Tyr187Cys* | -1.3 | 2.5 |
| 13 | 36909178 | G | A | p.Arg264Cys | -4.5 | 2.5 |
| 13 | 36909900 | G | T | p.Ala23Asp* | 1.8 | -3.5 |
| 13 | 36905729 | C | T | p.Cys272Tyr | 2.5 | -1.3 |
| 13 | 36905612 | A | G | p.Leu311Pro | -3.8 | -1.6 |
| 13 | 36905693 | G | T | p.Pro284Gln | -1.6 | -3.5 |
| 13 | 36878765 | C | T | P.Gly580Arg | -0.4 | -3.5 |
| 13 | 36888538 | C | A | p.Gly437Cys* | -0.4 | 2.5 |
Table 5: Hydrophobicity analysis of amino acid residues of Spartin (SPG20).
Splicing SNPs identified through HSF, SPICE & SPLICE- MAN
A total of 4 splicing variants were obtained through dataset which were subjected for analysis in Human Splice Finder, Spliceman and Spicev2.15 (Table 6).
| Chr | Position | Ref | Alt | Consequence | Spliceman | SPICE | HSF | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 distance | Ranking (L1) | SSF_ wt | SSF_ mut | MES _wt |
MES_m ut | SPiCE probab ility | SPiCEin ter_2thr |
WT score |
CV variatio n |
|||||
| 13 | 36886616 | T | C | c.1484-2A>G | 34699 | 66% | 65.51 | 0 | 3.11 | -4.93 | 1 | high | 73.84 | -39.21 |
| 13 | 36886374 | T | C | c.1643-2A>G | 36353 | 74% | 98.45 | 0 | 8.17 | 0.12 | 1 | high | 93.8 | -30.86 |
| 13 | 36905535 | C | T | c.1008+1G>A | 32671 | 55% | 87.2 | 0 | 8.63 | 0.12 | 1 | high | 87.99 | -30.5 |
| 13 | 36903498 | C | T | c.1164+1G>A | 36301 | 74% | 78.56 | 0 | 5.83 | -2.68 | 1 | high | 82.02 | -32.71 |
Table 6: Analysis of the effect of mutations on the splicing site through various bioinformatic tools of the Spartin protein (SPG20).
In case of Spliceman, c.1643-2A>G exhibited the high potential to disrupt the splicing site of spartin protein. In spice, score is either 0 (low) or 1 (high). The spice and human splice finder result also validated the potential of these variants to disrupt the normal splicing activity of the protein.
Discussion
Hereditary spastic paraplegia is a neurodegenerative syndrome associated with the progressive minor symptoms like cramps, unstable walk, feeling of stiffness in the legand falling occurfrequently (Giudice et al., 2014). The spartin is a mitochondrial protein mainly associated with troyer’s syndrome (autosomal recessive form of hereditary spastic paraplegia), plays a crucial role in the ubiquitination, protein folding, influx movement of Ca ions, as well as the degradation of the different proteins.The mitochondrial dysfunction can ultimately lead to axonal degeneration causing hereditary spastic paraplegia.
In the recent analysis, the data for variant analysis of spartin protein were obtained through gnomAD. After applying several filters and passing the data through a stringent analysis to study the effect of these mutations on the protein, total 9 nsSNPs were obtained. These nsSNPs were predicted to be causing highly deleterious effect on the normal structure, function or the stability of the protein. For SPG20, NM_001142296 was used to study the effect of mutation on the spartin protein.
The effect of mutations on the protein’s function is predominantly predicted based on the knowledge of the amino acid conservation, changes in the structure of protein at amino acid level, a huge list of annotations as well as the physicochemical analysis. In this study, the effect ofnsSNPson the protein’sstructurewaspredictedthrough I-Stable.
For splicing analysis, three tools named Spliceman, HSF and SPICE v2.0 were employed. After detailed analysis, 4 variants of SPG20 were obtained. The mutations analysis of all the four variants showed their capability to disrupt the normal splicing activity, thus may cause deleterious effect in the normal splicing process of the protein.
In this work, the hydrophobicity analysis was carried out based on kdHydrophobicity scale in combination with the visualization of hydrophobic amino acids properties in the Chimera. According to kyte and Doolittle scale, a value is prescribed to each amino acid based on either its hydrophilic or hydrophobic nature. The Chimera was further used to analyze the change in the hydrophobic properties of the amino acid’s residues upon substitution due to mutations.
To find novelty in the work, ClinVar was utilized. The server show results on the basis of immense data presented by different clinical labs, researchers, individual scientists or experts. In the present analysis, 3 missense mutations and 4 spicing mutations obtained were not have not been reported in any kind of databases or scientific work, thus can be considered asnovel.
Conclusion
Hereditary spastic paraplegia also known as HSP, primarily associated with the increasing spasticity as the complication arises in the lower limb area. In this work, the SNPs associated with the spartin (SPG20) were analyzed to check the impact of mutations (either deleterious or benign) on the protein. A list of bioinformatic tools were employed to study the predicted pathogenicity associated with the missense and splicing variants of Spartin. In the recent work, three missense and four splicing mutations reported in this work were found to be novel, in an aspect that they have been not reported in any previous research work. The SNPs found in this study can be analyzed further in experimental analysis conducted for hereditary spastic paraplegia.
Acknowledgement
None
Conflict of interest
None
References
- Salinas S (2008) Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. The Lancet Neurology 7: 1127-1138.
- Kasher PR (2009) Direct evidence for axonal transport defects in a novel mouse model of mutant spastin‐induced hereditary spasticparaplegia(HSP) and human HSP patients. Journal of neurochemistry 110: 34-44.
- Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. Journal of molecular biology 157: 105-132.
- Landrum MJ (2017) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic acids research 46: 1062-1067
- Laskowski RA (2016) Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Human molecular genetics 25: 927-935.
- Lim KH, Fairbrother WH (2012) Spliceman-a computational web server that predicts sequence variations in pre-mRNA splicing. Bioinformatics 28: 1031-1032.
- Lek M (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285-291
- Richards,S (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine 17: 405-423.
- Mooney S (2005) Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Briefings in bioinformatics 6: 44-56.
- Rentzsch P (2018) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic acids research 47:886-894.
- Yang J, Zhang Y (2015) Protein structure and function prediction using I-TASSER. Current protocols in bioinformatics 52: 5.8-5.8. 1.
- Berezin C (2004) ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20: 1322-1324.
- Pettersen E (2004) UCSF Chimera-a visualization system for exploratory research and analysis. Journal of computational chemistry 25: 1605-1612.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi