Research Article, J Clin Exp Onco Vol: 12 Issue: 4
Concordance and Deviations of the PDX Tumors from the Primary Tumors of NSCLC Patients: Effects of Murine Fibroblasts on Low Engraftment Rates
Jinseon Lee1*, Chae Hwa Seo2, Bo Kyung Kim1, Jung Hee Lee1, Jung Hee Kang1, Sung-Hyun Kim1, Minseob Cho3, Hong Kwan Kim1, Jong Ho Cho1, Yong Soo Choi1, Sumin Shin1, Young-Ae Choi1, Hyun Kuk Song2, Min Young Park2, Hye Yoon Jang2, Sanghyuk Lee4, Dae-Soon Son3, Jong-Eun Lee2**, Jhingook Kim1**
1Department of Thoracic and Cardiovascular Surgery, Sungkyunkwan University, Seoul, South Korea
2Division of Personal Genome, DNA Link Sequencing Lab, Seoul, South Korea
3Department of Data Science and Data Science Convergence Research Center, Hallym University, Chuncheon, South Korea
4Department of Life Science, Ewha Womans University, Seoul, South Korea
*Corresponding Author: Jinseon Lee,
Department of Thoracic and Cardiovascular
Surgery, Sungkyunkwan University, Seoul, South Korea
E-mail: jinseonl@gmail.com
*Corresponding Author: Jong-Eun Lee,
Division of Personal Genome, DNA Link Sequencing Lab, Seoul, South
Korea
E-mail: jonglee@dnalink.com
*Corresponding Author: Jhingook Kim,
Department of Thoracic and Cardiovascular Surgery, Sungkyunkwan
University, Seoul, South Korea
E-mail: jkimsmc@skku.edu
Received date: 04 August, 2023, Manuscript No. JCEOG-23-109106;
Editor Assigned date: 07 August, 2023, PreQC No. JCEOG-23-109106 (PQ);
Reviewed date: 21 August, 2023, QC No JCEOG-23-109106;
Revised date: 29 August, 2023, Manuscript No. JCEOG-23-109106 (R);
Published date: 05 September, 2023, 10.4172/2324-9110.1000361.
Citation: Lee J, Seo CH, Kim BK, Lee JH, Kang JH, et al. (2023) Concordance and Deviations of the PDX Tumors from the Primary Tumors of NSCLC Patients: Effects of Murine Fibroblasts on Low Engraftment Rates J Clin Exp Oncol 12:4.
Abstract
Patient-Derived Xenograft (PDX) models of primary lung cancer have been reported. However, varying engraftment rates and their underlying mechanisms for specific subtypes of lung cancer (adenocarcinoma, squamous cell carcinoma, and large cell neuroendocrine carcinoma) have not been studied. The authors prepared subcutaneous tumors grown in NOD Scid Gamma Mouse (NSG™) mice with primary tumors of lung cancer patients to develop lung cancer PDX models. Pathological features of the subcutaneous tumors were compared with those of the patients. One hundred 7teen lung cancer PDX models retaining the original pathologic features were obtained from 642 primary lung cancer patients. Nineteen PDX tumors and the corresponding patient tumors, representing three subtypes of lung cancer, were selected and analyzed with in-depth genomic and transcriptomic profiling. Results showed the PDX tumors retained most of the somatic and oncogenic mutations with limited levels of additional xenograft- specific mutations. Significant downregulation of the genes involved in hypoxia-associated angiogenesis was found compared with the corresponding human tumors. This downregulation was associated with murine fibroblasts in the PDX tumor microenvironment, which might be an important factor in low engraftment rates in primary lung cancer PDX models.
Keywords
Hypoxia; Angiogenesis; Murine fibroblast; Tumor microenvironment; Cell types; Primary lung cancer
Abbreviations
ANT: Adjacent Normal Tissue; CAF: Cancer-Associated Fibroblast; DEG: Differentially Expressed Gene; EBV: Epstein- Barr virus; EMT: Epithelial-Mesenchymal Transition; H&E: Hematoxylin and Eosin; IHC: Immuno Histo Chemistry; IRB: Institutional Review Board; LCNEC: Large Cell Neuroendocrine Carcinoma; LUAD: Lung Adenocarcinoma; LUSC: Lung Squamous Cell Carcinoma; NSCLC: Non-Small Cell Lung Cancer; PDX: Patient-Derived Xenograft; SCLC: Small Cell Lung Cancer; TME: Tumor Micro Environment; WES: Whole Exome Sequencing; WTS: Whole Transcriptome Sequencing; XALD: Xenograft-Associated Lymphoproliferative Disease.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide [1,2]. It is classified into two main histological categories: Non-Small Cell Lung Cancer (NSCLC) (85%) and Small Cell Lung Cancer (SCLC; 15%). NSCLCs are generally subcategorized into Lung Adenocarcinomas (LUADs), Squamous Cell Carcinomas (LUSCs), and Large Cell Neuroendocrine Carcinomas (LCNECs) [3,4]. With the aid of next generation sequencing technology, up to 60% of LUADs were shown to have a known oncogenic driver mutation as well as fusion or amplification in signaling pathways, which allowed the development of therapeutic agents that target specific molecular pathways. Despite this improvement in targeted therapy, secondary alterations in the downstream and or alternative pathways lead to acquired resistance and disease progression. On the contrary, therapeutic targets have not been clearly identified in LUSC and in LCNEC due to the rarity of these cancers [5,6].
Patient-Derived Xenograft (PDX) models have been developed using NSG™ mice as a means to preserve the histological structures in human tumors, even for limited passages [7]. These features allow the model to be used as a preclinical model for the development of target drugs against de novo resistance and as co-clinical models for the selection or combination of treatment regimens before clinical applications [8-13]. Cancer immunotherapy is one of the most promising approaches to refractory cancers and PDX tumors can be invaluable resources for preparing humanized PDX models for cancer immunotherapy using NSG mice infused with CD34 human hematopoietic stem cells [14]. These merits make the three major pathological subtypes of NSCLCs eligible for preparation of PDX models that can be used to find target drugs or immune therapeutics to control lung cancer.
In this study, the authors established 117 PDX models from 642 primary lung cancers. Representative models of LUAD, LUSC, and LCNEC subtypes were selected and the pathological, genomic, and transcriptomic patterns of PDX tumors were compared with those of corresponding patient tumors based on our previous studies using LUSC PDX models [15]. The factors responsible for the low success rate of the PDX models, particularly in the case of adenocarcinomas, were considered when attempting to improve the efficiency of generating PDX models, which could increase the application opportunities.
Materials and Methods
Tumor samples from patients with primary NSCLCs
Tumor samples were obtained from 642 primary NSCLC patients between September 2015 and December 2019. All patients provided signed informed consent. This study was approved by the IRB of Samsung Medical Center (2014-10-069, 2015-04-018, 2018-03-110). Table 1 shows the clinical characteristics of the NSCLC patients. Clinical features such as age, gender, preoperative chemotherapy treatments, smoking status, stage, tumor size, differentiation, recurrence, vascular invasion, perineural invasion, lymphatic invasion, visceral pleural invasion, and survival were obtained from medical records (Table 1).
| n=642 | PDX success | PDX failure | p-value | Odds Ratio (95% conf.) | |
|---|---|---|---|---|---|
| (n=117) | (n=525) | ||||
| Age (mean ± sd) | 65.2544±8.6052 | 62.9261±9.6130 | - | - | |
| Gender | Male (n=395) | 102 | 293 | <0.0001 | 5.3843 (3.0489, 9.5086) |
| Female(n=247) | 15 | 232 | |||
| Smoking status | Formerorcurrent (n=393) | 102 | 291 | <0.0001 | 5.4680 (3.0965, 9.6560) |
| Never (n=249) | 15 | 234 | - | - | |
| Preoperative chemotherapy | Yes (n=60) | 3 | 57 | 0.0053 | 0.2161 (0.0665, 0.7024) |
| No (n=582) | 114 | 468 | |||
| pTNM stage (7th) | I (n=284) | 28 | 256 | <0.0001 | 0.3306 (0.2091, 0.5226) |
| II (n=173) | 53 | 120 | - | - | |
| III (n=152) | 29 | 123 | - | - | |
| IV (n=33) | 7 | 26 | - | - | |
| Tumor size | <3cm (n=252) | 21 | 231 | <0.0001 | <3cm:0.2784(0.1684, 0.4602) |
| 3<<5 (n=259) | 46 | 213 | - | <5cm:0.2445(0.1580, 0.3781) | |
| 5<<7 (n=104) | 41 | 63 | - | - | |
| 7>(n=27) | 9 | 18 | - | - | |
| Differentiation | Well (n=12) | 1 | 11 | 0.0554 | 0.6021 (0.3905, 0.9282) |
| Moderate(n=418) | 66 | 352 | - | - | |
| Poor (n=179) | 42 | 137 | - | - | |
| UK (n=2) | 0 | 2 | - | - | |
| NA (n=31) | 8 | 23 | - | - | |
| Recurrence | Yes (n=256) | 51 | 205 | 0.3642 | 1.2062 (0.8043, 1.8090) |
| No (n=386) | 66 | 320 | - | ||
| Vascular invasion | Yes (n=52) | 16 | 36 | 0.0134 | 2.1728 (1.1603, 4.0689) |
| No (n=583) | 99 | 484 | - | - | |
| NA (n=7) | 2 | 5 | - | - | |
| Perineural invasion | Yes (n=31) | 10 | 21 | 0.0382 | 2.2417 (1.0259, 4.8984) |
| No (n=605) | 106 | 499 | - | - | |
| NA (n=6) | 1 | 5 | - | - | |
| Lymphatic invasion | Yes (n=214) | 48 | 166 | 0.0497 | 1.5096 (0.9989, 2.2812) |
| No (n=423) | 68 | 355 | - | - | |
| NA (n=5) | 1 | 4 | - | - | |
| 0.Visceralpleuralinvasion | PL0 (n=500) | 81 | 419 | 0.0005 | 0.3693 (0.2257, 0.6129) |
| PL1 (n=44) | 5 | 39 | - | - | |
| PL2 (n=55) | 17 | 38 | - | - | |
| PL3 (n=34) | 13 | 21 | - | - | |
| NA (n=9) | 1 | 8 | - | - | |
| Note: UK: Unknown; NA: Not Applicable. | |||||
Table 1: Clinical parameters affecting the success rate of primary lung cancer Patient-Derived Xenograft (PDX) models.
Establishment of primary lung cancer PDX models
To establish primary lung cancer PDX models, tumor samples from patients with primary lung cancer were subcutaneously implanted into the flanks of NSG mice (Jackson Laboratory, Sacramento, CA, USA). The size of a mouse subcutaneous tumor was measured with a caliper twice a week until it reached 60 mm3 in volume. Tumor volumes were calculated as 0.5 x length x width2. The mice were sacrificed when the tumor size reached 600-800 mm3. Then, the subcutaneous tumors were surgically harvested for subsequent procedures. Expansion of the tumor tissues was carried out with three passages. Formalin-fixed paraffin-embedded samples were prepared for pathologic examination. Short tandem repeat analysis was carried out for direct identification of mouse subcutaneous tumors, and next generation sequencing analysis was subsequently performed. All animals were cared for and treated following an animal protocol that had been approved by the CHA Advanced Research Institute and Biomedical Research Institute at Seoul National University Hospital.
Whole Exome Sequencing (WES)
Three micrograms of genomic DNA were used to establish DNA libraries. Using an Agilent SureSelect Human All Exon V3 kit (Agilent Technologies, Santa Clara, CA, USA), target enrichment was performed, following the manufacturer’s instructions, to generate exome sequencing libraries. Exon capturing was then followed using an Agilent SureSelect 50Mb system. Paired-end DNA sequences were obtained with the Illumina sequencing system HiSeq 2000 (Illumina Inc., San Diego, CA, USA). The sequenced reads were aligned to the human genome information from the University of California Santa Cruz hg19. MuTect, VarScan 2, and the GATK Somatic Indel Detector were used to identify somatic mutations, and these mutations were later verified through Sanger sequencing. Significantly mutated genes were identified with MutSigCV, and functional enrichment of the somatic mutations was assessed with Metacore (GeneGo Inc., St. Joseph, MI, USA). The GRCh37 reference was used for sequencing data analysis.
Whole Transcriptome Sequencing (WTS)
mRNA libraries (insert size of ~300 bp) were prepared with a TruSeq RNA Library Preparation Kit v2 (Illumina Inc., San Diego, CA, USA). A total of 1 μg of RNA from each case sample was used to create the library. The samples were subjected to 101-bp paired- end sequencing using the Illumina sequencing system HiSeq 2000. Library preparation and sequencing were performed at DNA Link, Inc.
Differentially Expressed Gene (DEG) analysis
Genes with at least one sample indicating a sequencing read count of at least two for each were initially screened. Differential expression of each gene was analyzed by calculating the log2 (fold change) value for the gene expression of PDX tumors relative to that of patient tumors, or the Adjacent Normal Tissue (ANT), depending on the purpose of the analysis. The genes with a false discovery rate <0.05 and a |log2 (fold change)|>2 were selected as upregulated or downregulated genes, respectively.
Pathological analysis
Formalin-fixed paraffin-embedded tumor tissues from primary lung cancer patients and corresponding subcutaneous tumors were freshly cut into slices of 4 μm. Following the manufacturer’s instructions, Hematoxylin and Eosin (H&E) staining was performed using Symphony (Ventana Medical Systems, Inc., Roche, Basel, Switzerland). Immuno Histochemical (IHC) staining for CK5, p63, Thyroid Transcription Factor (TTF-1), pan-cytokeratin, or CD56 was performed on a single representative block with the following procedures. Deparaffinized slides were treated with citrate buffer (pH 6.0) for antigen retrieval. Next, the primary antibody was incubated with the Dako Antibody Diluent (S3022, Dako, Agilent Technologies, Inc., Santa Clara, CA, USA) and then with Dako REAL EnVision Detection System (K5007, Dako, Agilent Technologies, Inc., Santa Clara, CA, USA). The images obtained from H&E and IHC staining were analyzed with a Scan Scope® XT scanner (Aperio, Leica Biosystems, Newcastle, UK). Antibody sources and dilution factors are shown in Table S1. LUAD was determined by TTF-1+ CK5- CD56- p63+/-, LUSC by TTF-1- CK5+ CD56- p63+, and LCNEC by TTF-1+/- CK5- CD56+ p63-. Pathologically unmatched subcutaneous tumors were further categorized into Xenograft- Associated Lymphoproliferative Disease (XALD) or epithelial tumor not identical to that of the patient.
Statistical analyses
The association between PDX model success rates and patient characteristics was investigated with a chi-squared test. The Cox proportional hazards model was used for multiple analyses of clinically significant prognostic variables. Statistical analyses were carried out with R software for Windows version 4.2.1 (The R Foundation, St. Miami, FL, USA). The odds ratio and its confidence interval were calculated with the “epitools” package of R software. All p-values were two-sided and less than 0.05 was considered statistically significant.
Results
Establishment of primary lung cancer PDX models
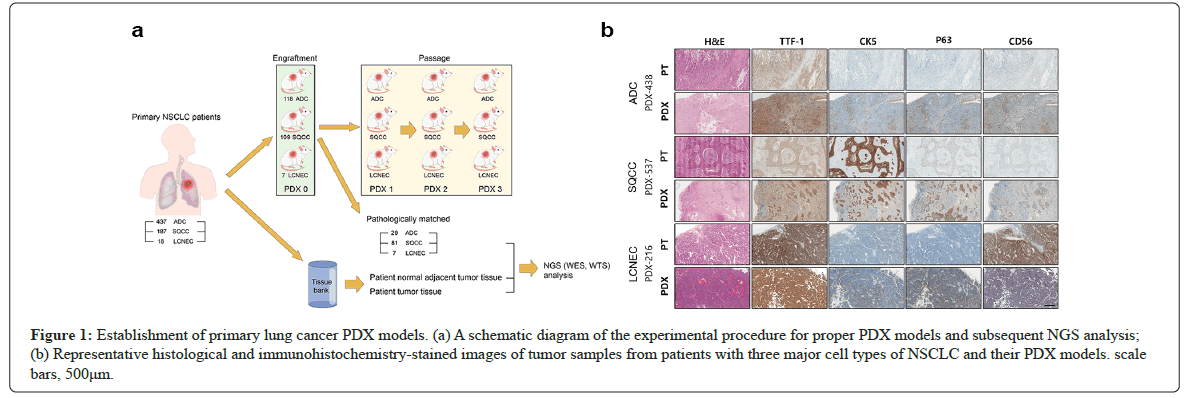
A total of 642 patients with LUADs, LUSCs, and LCNECs were enrolled in the study between September 2015 and December 2019. Tumor samples from these patients were grafted into NSG mice to establish primary lung cancer PDX models for each cell type. The subcutaneous tumor generation rates were 27.0% in 118 cases from 437 LUAD tumor samples, 58.3% in 109 cases from 187 LUSC tumor samples, and 38.9% in 7 cases from 18 LCNEC tumor samples, as shown in Figure 1a.
Figure 1: Establishment of primary lung cancer PDX models. (a) A schematic diagram of the experimental procedure for proper PDX models and subsequent NGS analysis; (b) Representative histological and immunohistochemistry-stained images of tumor samples from patients with three major cell types of NSCLC and their PDX models. scale bars, 500μm.
To validate pathological concordance with the patient tumors, cell types were examined with antibodies against TTF-1, CK5, p63, and CD56 proteins to differentiate human LUAD, LUSC, and LCNEC. For each cell type, representative Immuno Histo Chemistry (IHC) images of PDX tumors and the corresponding patient tumors are shown in Figure 1b. Subcutaneous tumors that did not match the cell types of the patient tumors were further classified into either epithelial or non-epithelial (XALD) cell types (Table 2a).
| Lung Cancer cell type | Number of engraftment attempts | Number of subcutaneous tumor generated(% attempts) | |||
|---|---|---|---|---|---|
| Pathologically | |||||
| matched (n=117) | |||||
| unmatched (n=117) | |||||
| Model (% subcutaneous tumor) | Epithelial tumor(% unmatched) | Non-epithelial tumor(XALD:% unmatched) | |||
| LUAD | 437 | 118(27.0%) | 29(24.6%) | 20/89(22.5%) | 69/89(77.5%) |
| LUSC | 187 | 109(58.3%) | 81(74.3%) | 8/28(28.6%) | 20/28(71.4%) |
| LCNEC | 18 | 7(38.9%) | 7(100%) | 0 | 0 |
Table 2a: Engraftment of human tumors and statistics for primary lung cancer Patient-Derived Xenograft (PDX) models, statistics for overall PDX model success rates.
The order of efficiency in subcutaneous tumor generation was LUSC, LCNEC, and LUAD; however, except for the pathologically irrelevant tumor, the PDX model success rate was in the order of LCNEC, LUSC, and LUAD. When LUAD was subtyped further into five categories based on histological characteristics, the PDX model success rates were highest in the solid subtype and lowest in the papillary subtype. LUAD had the lowest PDX model success rate (6.6% on average), which was only between 1/6 and 1/7 of those for LUSC or LCNEC (43.33% and 38.9%, respectively; (Table 2b). Tumors from patients with an advanced stage of NSCLC tended to have higher PDX model success rates (Table 2c).
| Tumor cell type | A | B | C | |
|---|---|---|---|---|
| LUAD | 437 | 29 | 6.60% | |
| Solid | 113 | 14 | 12.40% | |
| Lepidic | 16 | 1 | 6.30% | |
| Micropapillary | 66 | 3 | 4.50% | |
| Acinar | 122 | 4 | 3.30% | |
| Papillary | 82 | 1 | 1.20% | |
| others | 38 | 6 | 15.80% | |
| LUSC | - | 187 | 81 | 43.30% |
| LCNEC | - | 18 | 7 | 38.90% |
Table 2b: Engraftment of human tumors and statistics for primary lung cancer PDX models, PDX model success rates for the subtypes of primary lung cancer.
| Stage | LUAD | LUSC | LCNEC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | A | B | C | |
| I | 219 | 7 | 3.20% | 58 | 20 | 34.50% | 7 | 1 | 14.30% |
| II | 85 | 9 | 10.60% | 81 | 42 | 51.90% | 7 | 2 | 28.60% |
| III | 102 | 8 | 7.80% | 46 | 17 | 37.00% | 4 | 4 | 100% |
| IV | 31 | 5 | 16.10% | 2 | 2 | 100% | - | - | - |
| Total | 437 | 29 | 6.60% | 187 | 81 | 43.30% | 18 | 7 | 38.90% |
Table 2c: Engraftment of human tumors and statistics for primary lung cancer PDX models, PDX success rates depending on the tumor stage.
Among the pathologically irrelevant subcutaneous tumors, the ratio between the epithelial vs non-epithelial (XALD) cases were 22.5% vs 77.5% in LUADs, and 28.6% vs 71.4% in LUSCs, indicating no major difference between the two cell types. A total of 29 LUAD PDX tumors, 81 LUSC PDX tumors, and 7 LCNEC PDX tumors displayed the same pathology as the patient tumors. Clinical parameters of 642 patients were analyzed to select 12 parameters that influenced the engraftment rates (Table 1). The odds ratios were highest in the patients who were not treated with preoperative chemotherapy and second highest in males. In males, visceral pleural invasion and pStage III or lower were next highest among the advanced stage factors, while in females, tumor size and lymphatic invasion were the next highest.
Concordance of somatic mutations in OncoPanel genes in PDX tumors
Seven LUAD cases, eight LUSC cases, and four LCNEC cases were selected from 117 pathologically relevant PDX models to compare the somatic mutations between PDX and patient tumor pairs. OncoPanel genes were analyzed to test whether the driver mutations of the patient tumors were retained in the PDX tumors (Figure S1) [16].
Somatic mutations that were absent in the ANT were classified into three groups; that is, those common to PDX models and patient tumors, those found only in patient tumors, and those only in PDX models. In general, most somatic mutations were commonly seen in both PDX models and patient tumors, but a significant number of patient- or PDX-specific somatic mutations that were different depending on the patients and the cell types were also found (Table 3).
| Tumor cell type | PDX model ID | Patient | PDX models | |
|---|---|---|---|---|
| Patient-specific | Common somatic | PDX model-specific | ||
| mutations | mutations | mutations | ||
| ADC | 13 | ERCC5, HNF1A, STAG2 | CDKN2A, CUX1, FLT3, NF1, NOTCH1, PBRM1, POLE, TP53, TSC2 | DOCK8, ERCC4, MAP2K2 |
| 41 | none | none | TP53, NF1, ESR1, POLE, SLX4, CCNE1, PIK3R1, TRIM37 | |
| 48 | none | MET, POT1 | TP53, JAK2, KCNQ1 | |
| 75 | SDHA, CARD11, FAH, PARK2 | ABCB11, BCL2, BRCA2, ENG, SLX4, TP53 | NF1, NOTCH2 | |
| 81 | none | BRCA1, CDH4, ERCC3, JAK2, KEAP1, MGA, MYB, TP53 | FOXL2 | |
| 29 | none | PBRM1 | CHEK2, FAT1 | |
| 216 | TDG, CRTC3 | FGFR4, NOTCH1, REL, TP53 | none | |
| SQCC | 231 | TDG | COL7A1, DCLRE1C, FLT1, FLT3, GNAS, KEAP1, MET, NF2, NR4A3, NUP214, STK11, TP53 | APC, SMO, KDM5C |
| 244 | ERCC5, FANCC, NTRK1 | ABCB11, CUX1, HLEQ, PIK3CA, SETBP1, SMARCA4, TET1, TP53 | EP300, ARHGAP35, IL7R | |
| 295 | none | none | TP53, BCOR, ETV1, FLCN, TFE3, USP8, WT1 | |
| 363 | NRG1 | ASXL1, BRCA2, NEIL3, TP53 | CHEK2, SDHC, GATA2, POLQ, SOX2 | |
| 367 | ATM, KRAS, BARD1, NRG1, POT1, ARHGEF12, BRCC3, EXO1, FOXL2, KIF1B | RBM10 | ERBB4, ARID1A, PDGFRA, TDG | |
| 369 | SDHC | ALK, ATM, CREBBP, GBA, KRAS, PDGFRB, TP53 | none | |
| 415 | none | APC, CDKN2A, CREBBP, DNMT3A, NFE2L2, TP53 | FLT3, SDHA, GNAQ, CYLD | |
| 416 | none | ATR, ERBB4, HFE, MITF, POLD1, RB1, SMC3, SMO, TP53 | NOTCH2, SDHA, ESR1 | |
| LCNEC | 438 | SDHC, PTEN | EGFR, GNAS | TP53, DOCK8, ABL1, MDM2 |
| 451 | FAT1, MAX | ATM, CDK8. ERBB4, FGFR4, FLT3, NOTCH2, PDGFRB, PHOX2B, PIK3CA, QKI, TP53 | EP300, EXT2, JAK1 | |
| 482 | BARD1 | GNAQ, HELQ, KRAS, PIK3CA | DIS3L2 | |
| 508 | COL7A1, GNAQ, KDR, MTOR | CHEK2, MECOM, NF1, NOTCH1, RB1, SDHA, TP53 | HIST1H3B, KAT6B, RIT1 | |
Table 3: Somatic mutations in OncoPanel genes in tumors of PDX models and patients.
Most somatic mutations found in the patient tumors were retained in the PDX tumors, suggesting that the PDX models could be utilized in preclinical studies. Nonetheless, as not all mutations in the patient tumors were retained in the PDX models; e.g., Kirsten Rat Sarcoma Virus (KRAS) mutation negative in LUAD PDX-367 in Table 3, retention of the mutations of interest should be confirmed in the early stages of use of the PDX model.
Pre-existing mutations might disappear or additional mutations might appear in PDX tumors, for the following reasons: 1) The patient tumor fragments used for pathological examination did not have an identical genetic makeup from those for PDX models due to intra-tumoral heterogeneity, and or 2) de novo mutations might occur during multiple passage processes for the expansion of tumor tissues. For example, new PDX-specific mutations were found in TP53 in four independent models and in Neuro Fibromatosis Type 1(NF1), Neurogenic Locus Notch Homolog Protein 2 (NOTCH2), Checkpoint Kinase 2 (CHEK2), EP300, Succinate Dehydrogenase Complex Flavoprotein Subunit A (SDHA), Dedicator of Cytokinesis Protein 8 (DOCK8), and Estrogen Receptor 1 (ESR1) in more than two independent models.
Deviations in gene expression in PDX tumors
Despite most somatic mutations being commonly found in both the patient and PDX tumors, existence of the patient or PDX-specific mutations suggested possible discrepancies in gene expression. To address this question, 460 genes in OncoPanel were analyzed for gene expression, and those with |log2 (fold change)| >2 with respect to the ANT were selected as DEGs in patient or PDX tumors (Figure S1 and Table 4).
| Tumor cell type |
|
Upregulated | Downregulated | |
|---|---|---|---|---|
| ADC | DP-013 | Patient | None | None |
| PDX | None | None | ||
| DP-041 | Patient | None | None | |
| PDX | (7) HIST1H3B; (4) EXO1, MYB;(3) BCL2, BRCA1, CARD11, CDKN2A, DCLRE1C; (2) BARD1, COL7A1, NEIL3, POLD1 | (-6) NR4A3; (-5) ENG, ETV1; (-4) PDGFRA; (-3) GNAQ, MECOM; (-2) CUX1, MET, MITF, NFE2L2, NOTCH2, QKI | ||
| DP-048 | Patient | (4) CDKN2A; (3) HIST1H3B | (-4) FGFR4 | |
| PDX | (5) HIST1H3B; (4) CDKN2A, MET; (3) BRCA1, COL7A1, POLQ; (2) NF2, POT1, TRIM37 | (-5) PIK3R1, (-4) PDGFRB, (-2) FANCC, IL7R | ||
| DP-075 | Patient | (5) COL7A1; (3) HIST1H3B; (2) FAT1, KRAS, POLQ | (-3) ENG | |
| PDX | (5) COL7A1, HIST1H3B; (3) KRAS, POLQ; (2) FAT1, SOX2 | (-5) DOCK8, ETV1, PTEN; (-4) ENG; (-3) FLT1, GATA2, GNAQ, IL7R; (-2) ESR1, FGFR4, JAK2, MGA | ||
| DP-08 1 | Patient | (4) COL7A1; (3) HIST1H3B, SMO | None | |
| PDX | (5) COL7A1; (4) HIST1H3B, NGR1; (3) CARD11, EXO1, POLQ; (2) BRCA1, MET | (-4) ETV1; (-3) ENG, IL7R, JAK2, KDR, MITF, PDGFRA, PDGFRB; (-2) DOCK8, GATA2, GNAQ, PIK3R1, QKI | ||
| SP-029 | Patient | (3) HIST1H3B, TSC1; (2) BCOR | (-3) KDR | |
| PDX | (3) BCOR, POLE; (2) ARID1A, DNMT3A, ETV1, HIST1H3B | (-5) FLT1; (-4) IL7R, JAK2, QKI; (-3) ETV1, MET, MITF, PIK3R1; (-2) ENG, MECOM, REL | ||
| SP-216 | Patient | (5) CDKN2A, HIST1H3B; (4) EXO1; (3) POLQ; (2) BARD1, BRCA1, POLE, TRIM37 | (-5) DOCK8; (-4) IL7R, JAK2, QKI; (-3) ETV1, MET, MITF, PIK3R1; (-2) ENG, MECOM, REL | |
| PDX | (5) CDKN2A; (4) EXO1, HIST1H3B; (3) POLQ; (2) BARD1, BRCA1, POLE, BCL2, BCOR, BRCA2, BRCC3, HFE, MYB, STAG2, TET1 | (-5) DOCK8, QKI; (-4) FGFR4, JAK2, PDGFRA; (-3) ENG, IL7R, MET, PIK3R1, FAH, FLT1, PDGFRB; (-2) ETV1, REL, KCNQ1, KDR, PTEN, SMO | ||
| SQCC | SP-231 | Patient | (4) COL7A1 | (-3) NR4A3; (-2) EGFR, MECOM |
| PDX | (4) HIST1H3B; (3) CDKN2A, NRG1, POLD1, TDG; (2) BRCA1, BRCA2, CHEK2, FAT1, KIF1B, MTOR, POLE, SMO | (-5) ENG, IL7R, KDR, MECOM, PDGFRA; (-4) APC, PIK3R1; (-3) CYLD, GNAQ; (-2) ESR1, GATA2, NOTCH1, NR4A3, QKI | ||
| SP-244 | Patient | (3) HIST1H3B, KDR; (2) BARD1 | (-2) ENG, GATA2, SETBP1 | |
| PDX | (3) BARD1, HIST1H3B; (2) TET1 | (-5) ENG, ETV1, PDGFRA; (-4) FLT1, IL7R; (-3) DOCK8, GATA2, QKI; (-2) NOTCH1, PIK3R1 | ||
| SP-295 | Patient | None | None | |
| PDX | (6) NRG1; (3) FAT1, POLQ; (2) ATR, BRCA1, BRCA2, CDK8, CHEK2, HIST1H3B, KEAP1, SMARCA4 | (-5) NR4A3; (-4) GATA2; (-3) ETV1, FGFR4, WT1; (-2) DOCK8, ALK, ESR1, PDGFRB | ||
| SP-363 | Patient | (6) CDKN2A, HIST1H3B; (3) BRCA1, CHEK2; (2) NEIL3, SOX2 | (-3) ENG, NR4A3, PDGFRA, PDGFRB; (-2) DOCK8, JAK2, KDR | |
| PDX | (7) HIST1H3B; (4) BRCA1, CDKN2A; (3) NEIL3; (2) CHEK2, SOX2, BRCC3, DNMT3A, GBA, HFE, SMO, | (-5) PDGFRA; (-4) FLT1; (-3) DOCK8, IL7R, MECOM; (-2) ENG, FGFR4, JAK2, NOTCH1, RB1, STAG2 | ||
| SP-367 | Patient | (3) HIST1H3B | (-3) GATA2 | |
| PDX | (5) HIST1H3B; (4) BCL2, POLQ; (3) BRCA1, CARD11, MYB; (2) BARD1, BRCA2, CDKN2A, POLE | (-5) IL7R, MECOM; (-4) PDGFRA; (-3) MITF, PIK3R1; (-2) FLT1, NOTCH2 | ||
| SP-369 | Patient | (4) HIST1H3B, SOX2; (3) CARD11, COL7A1; (2) EGFR, KEAP1 | (-4) NR4A3; (-3) KDR, PTEN, REL; (-2) DOCK8, ENG, FLT1, IL7R, MITF | |
| PDX | (6) SOX2; (4) IST1H3B; (3) CARD11, COL7A1, KEAP1, POLQ, SMO; (2) EGFR,, BRCA1, MAP2K2, SDHA | (-6) PTEN; (-4) ENG, IL7R, PDGFRA; (-3) DOCK8, FGFR4; (-2) MITF, REL | ||
| SP-415 | Patient | None | (-3) NR4A3 | |
| PDX | (6) MYB; (5) EXO1; (3) BRCA1, CDKN2A, POLQ; (2) ATR, CHEK2, EP300, HIST1H3B, KRAS, POLE, SMARCA4 | (-5) FAT1, FLT1, MECOM; (-4) ENG, KDR, PDGFRA; (-3) JAK1, PIK3R1; (-2) CYLD, PTEN | ||
| SP-416 | Patient | (5) COL7A1, HIST1H3B, SOX2; (4) CDKN2A; (3) CHEK2, EXO1, SMO; (2) ATR | (-4) GATA2; (-3) ENG, IL7R; (-2) ERBB4, KDR | |
| PDX | None | None | ||
| LCNEC | SP-438 | Patient | (6) COL7A1; (4) HIST1H3B; (3) CARD11, MET; (2) EGFR | (-2) MECOM, SETBP1 |
| PDX | (4) HIST1H3B, MET; (3) COL7A1; (2) CARD11, EGFR | (-4) DOCK8, IL7R; (-3) ENG, KCNQ1, PIK3R1; (-2) ERBB4, GATA2, NOTCH1, QKI | ||
| SP-451 | Patient | (6) COL7A1; (4) HIST1H3B; (3) FAT1, SMO, STAG2; (2) BRCA1 | (-3) FGFR4, GATA2, KDR, NR4A3; (-2) ERBB4 | |
| PDX | (6) COL7A1; (5) HIST1H3B; (4) BRCA1, SMO; (3) NRG1; (2) ATR, CARD11, CHEK2, EGFR, FAT1, MYB, TET1 | (-4) PDGFRB; (-3) DOCK8, ENG, ETV1, GATA2, PDGFRA; (-2) IL7R | ||
| SP-482 | Patient | (3) HIST1H3B, MDM2; (2) GNAS, SOX2 | (-3) FGFR4; (-2) GATA2, NR4A3, SETBP1 | |
| PDX | (5) HIST1H3B; (3) MDM2, SOX2, EXO1, NEIL3; (2) BCL2, BRCA1, BRCC3, CCNE1, DNMT3A, EGFR, GNAS, POLD1, POLE, POLQ, SMO, TP53 | (-5) MECOM, PDGFRA; (-4) DOCK8, IL7R; (-3) JAK1; (-2) ESR1, NFE2L2, PIK3R1, QKI, RB1 | ||
| SP-508 | Patient | None | None | |
| PDX | None | None | ||
Table 4: Differentially expressed oncopanel genes in tumors of PDX models and patients with respect to ANTs The numbers in parenthesis indicate the value of log2 (fold change).
First, there was a trend for more genes to be either up or downregulated in PDX tumors than in patient tumor tissues. HIST1H3B was significantly upregulated in both patient and PDX tumors with little exception, which might indicate a cancer-specific phenomenon, supported by the fact that it is engaged in wrapping newly synthesized DNA as a core component of the nucleosome [17]. On the other hand, ENG was downregulated in both patient and PDX tumors; particularly, in more than 60% of the cases, ENG and Platelet Derived Growth Factor Receptor Alpha (PDGFRA) were downregulated together. Those genes related to wound healing (COL7A1), or tumor cell proliferation (HIST1H3B, Breast Cancer Gene1 (BRCA1), Cyclin-Dependent Kinase Inhibitor 2A (CDKN2A), and POLQ) were upregulated, and those involved in angiogenesis, hypoxia, and connective tissue remodeling (Endoglin (ENG), Platelet Derived Growth Factor Receptor Alpha (PDGFRA), GATA Binding Protein 2 (GATA2), and Kinase Insert Domain Receptor (KDR)) were downregulated [18-23].
It is known that extracellular matrix remodeling or angiogenesis is induced by hypoxia, and the oxygen concentration in the tumor is regulated by the master regulator HIF-1a [24]. Therefore, it was necessary to investigate the association between HIF-1a expression levels with hypoxia or angiogenesis in PDX tumors. The HIF- 1a expression in PDX tumors, patient tumors, and ANTs was comparatively analyzed for hypoxia (Table S2). HIF-1a expression levels in LUAD and LUSC PDX tumors were higher than ANTs, but lower than the patient tumors, at the level of 62%~75%. In other words, hypoxia occurred in most PDX tumors at a relatively lower level than the patient tumors, which was similar to the above observation that the four genes in OncoPanel (ENG, PDGFRA, GATA2 and KDR) were involved in hypoxia and angiogenesis were downregulated in PDX tumors compared with patient tumors.
Downregulated expression of hypoxia- and angiogenesisrelated genes in PDX tumors
To understand whether the lower expression of hypoxia- or angiogenesis-associated genes in PDX tumors was a consistent phenomenon, data from Gene Set Enrichment Analysis and the signature genes for hypoxia or angiogenesis in recent reports were consolidated to create expanded gene sets (Tables S3 and S4). Among these two gene sets, DEGs with |log2 (fold change)|>2 were selected from the DEG mother database described in the Methods section. The selected genes were associated with the TME component cells in which those genes were generally expressed; i.e., epithelial tumor cells, murine Cancer-Associated Fibroblasts (CAF), Endothelial cells, and Immune cells of PDX tumors (Table 5).
In comparison to the patient tumors, hypoxia-related genes that were downregulated in PDX tumors commonly to LUAD and LUSC, to LUAD and LCNEC, and to LUSC and LCNEC are listed in Table 5a. Twenty genes were mainly expressed in fibroblasts, and four genes in endothelial cells. Likewise, angiogenesis-related genes that were downregulated in PDX tumors commonly to LUAD and LUSC, to LUAD and LCNEC, and to LUSC and LCNEC are listed in Table 5b. Thirty-five genes were mainly expressed in fibroblasts and 28 genes were mainly expressed in endothelial cells. C-X-C Chemokine Receptor Type 4 (CXCR4), Versican (VCAN), Familial Adenomatous Polyposis (FAP), Fibronectin 1(FN1), FOS, Transforming Growth Factor Beta-3 (TGFB3), and C-C Motif Chemokine Ligand 2 (CCL2) were expressed in fibroblasts, and Fms Related Receptor Tyrosine Kinase 1(FLT1), TEK, Angiopoietin 2 (ANGPT2), and Secreted Phospho Protein 1(SPP1) were expressed in endothelial cells that belonged to both hypoxia- and angiogenesis gene sets.
| TME cell type | Downregulated | Upregulated | ||||
|---|---|---|---|---|---|---|
| ASL | AS | SL | AL | S early | L late | |
| Fibroblast | BGN, CXCR4, COL5A1, DCN, DUSP1, OLFML2B, TAGLN, VCAN | - | COL1A1, COL6A3, CTHRC1, FAP, FN1, FOS, LOX, PLIN2, RRAGD, TGFB3 | ACKR3, CCL2 | CDKN3, TPBG | - |
| Endothelial cells | FLT1 | TEK | ANGPT2, SPP1 | - | FOSL1, GRIN2D | |
| Others (None of the above, or ubiquitous) | - | - | - | - | GPRIN1, MFI2, ANLN | ATP11, BASP1, CP, ITPR3 |
Table 5a: Differentially expressed hypoxia- and angiogenesis-related genes in tumors of PDX models with respect to patients and their association with TME component cell types, Hypoxia-related genes.
| TME cell type | Downregulated | Upregulated | ||||
|---|---|---|---|---|---|---|
| ASL | AS | SL | AL | S early | L late | |
| Tumor cells | - | TNFRSF1B | AXL, SLC34A2 | - | AURKA, BIRC5, E2F1, ECT2, UBE2T | PRODH |
| Fibroblast | COL3A1, COL15A1, CXCL12, | CCL19, ITGA11, LRRC32, MMP19 | COL5A2, FAP, FN1, FOS, FSTL1, MEF2C, MMP11, TGFB3, THBS1, TIMP2 | CCL2, CXCL14, JAM3 | AURKB, JAG2, MYBL2, SOX2 | - |
| CXCR4, DDR2, GREM1, ITGB2, LDB2, LUM, MMP9, PDGFRA, PDGFRB, POSTN, PRELP, SERPINF1, STAB1, TIMP3, VCAN | ||||||
| Endothelial cells | A2M, ACVRL1, FLT1, OLR1, PLVAP, SPRACL1, VWF | APLNR, CD93, CLEC14A, | ANGPT2, ESAM, SPP1 | CALCRL, RGS5, THY1 | ETV4, HMGA1, SLC7A5 | ID1 |
| CNRIP1, ECSCR, EDNRB, ELTD1, FLT4, GIMAP8, GPR124, ROBO4, RUNX1T1, TEK, THBD, TIE1 | ||||||
| Immune cells | - | - | TNFSF12 | - | - | - |
Table 5b: Differentially expressed hypoxia- and angiogenesis-related genes in tumors of PDX models with respect to patients and their association with TME component cell types, Angiogenesis-related genes.
Most hypoxia and angiogenesis-related genes that were mainly expressed in fibroblasts were downregulated. Therefore, low HIF- 1a expression as well as low levels of expression in hypoxia- and angiogenesis-associated gene sets in PDX tumors might be caused by the murine TME, specifically murine fibroblasts. Unlike LUAD or late stage LUSC PDX tumors, early stage LUSC PDX tumors had relatively upregulated genes, compared to the patient tumors: For the hypoxia-related genes, Cyclin Dependent Kinase Inhibitor 3 (CDKN3) and Trophoblast Glycoprotein (TPBG) were expressed in fibroblasts, and FOSL1 and Glutamate Ionotropic Receptor NMDA type subunit 2D (GRIN2D) were expressed in endothelial cells. For angiogenesis-related genes, Aurora A Kinase (AURKA), Baculo Viral IAP Repeat Containing 5 (BIRC5), E2F Transcription Factor 1 (E2F1), Epithelial Cell Transforming 2 (ECT2), and Ubiquitin-Conjugating Enzyme E2T (UBE2T) were expressed in tumor cells, AURKB, Jagged Canonical Notch Ligand 2 (JAG2),
Mannose-Binding Lectin 2 (MBL2), SOX2 in fibroblasts, and ETS Translocation Variant 4 (ETV4), High Mobility Group A1(HMGA1), Solute Carrier Family 7 Member 5 (SLC7A5) in endothelial cells. Relatively high PDX engraftment rates in LUSCs appeared partly associated with upregulation of the hypoxia- and angiogenesisrelated genes. On the contrary, PDX tumors of late stage LCNEC cell types had one upregulated gene (i.e., Proline Oxidase Dehydrogenase PRODH) in tumor cells and one upregulated gene (i.e., Inhibitor of Differentiation1 (ID1)) in endothelial cells.
Discussion
PDX animal models are generated by grafting patient-derived tumors to immune deficient mice, which reconstitutes the tumors with pathological relevance to the original patients. It is necessary to timely generate the PDX models effectively when studies with animal models are needed for specific cancer patients. In spite of the effort paid to establish PDX models, not much in-depth discussion has occurred on the key factors that determine the PDX engraftment rates. Molecular genetic deviations from the original patient tumors, such as somatic mutations and altered gene expression , may produce pathologically irrelevant subcutaneous tumors, causing transitions to different epithelial cell types or to non-epithelial tumors (XALD) (70% and 30%, respectively in Table 2), resulting in low engraftment rates [25,26].
In this study, by analyzing the differences in somatic mutations and gene expression between the PDX tumors and the tumor of primary NSCLC patients, we aimed to understand the reason for low PDX engraftment rates in NSCLC at a molecular level. To this end, the authors took a retrospective data-collection and analyses approach, rather than hypothesis-generation and test approach. Typically, tumors generate local hypoxia as they grow, and hypoxiarelated genes, including HIF-1a, are activated. A variety of adaptive autophagic responses are initiated, such as Epithelial-Mesenchymal Transition (EMT) of tumor cells and dedifferentiation into cancer stem cells [27-31].
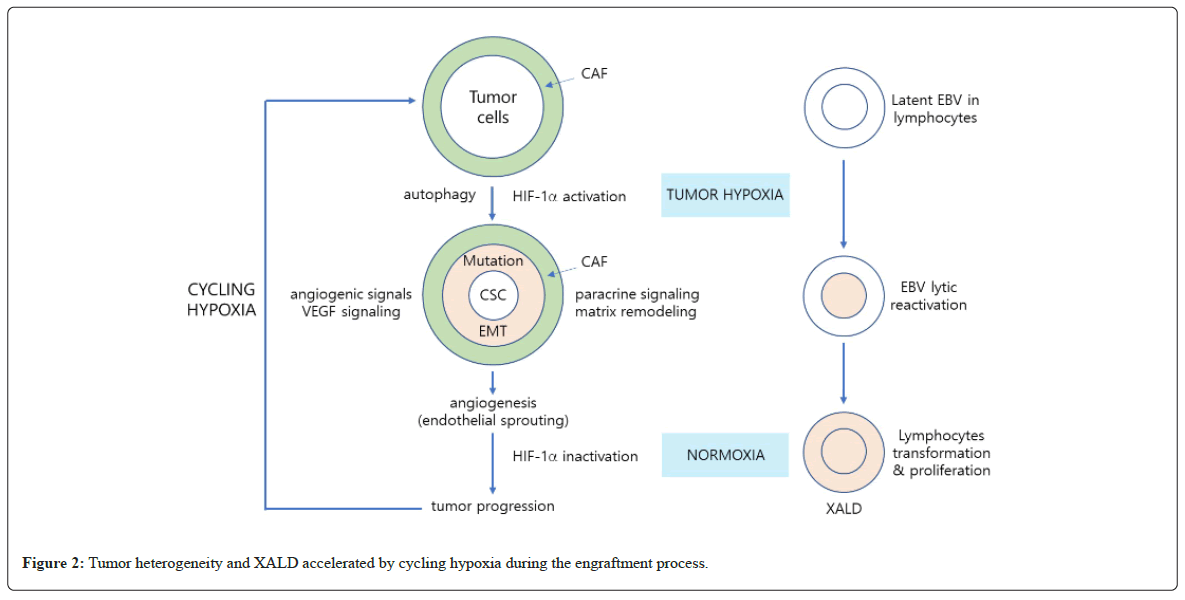
Tumor hypoxia increases mutation burdens, particularly enriching driver mutations in Tumor Protein (TP53), MYC, and Phosphatase and Tensin Homolog deleted on Chromosome 10 (PTEN10) [32-35]. At the same time, a local oxygen gradient appears in CAF in the TME that aids tumor subclonal evolution, resulting in further intratumor heterogeneity [36]. Through aberrant paracrine signaling and matrix remodeling, the niches necessary to maintain cancer stem cells are generated and the angiogenic signals stimulated by hypoxia support endothelial sprouting and tumor growth as well [37-45]. Angiogenesis reoxygenates tumors and HIF-1a becomes inactivated, but as the tumor grows local hypoxia is re-generated [46,47]. Through this cycling hypoxia, further clonal evolution facilitates an even more complex genomic situation [48-50] (Figure 2).
When grafted to NSG mice, most tumors form internal hypoxia at a relatively early stage, as supported by increased TP53 somatic mutations in a PDX-specific manner (Table S2, Table 3). During serial passaging to expand the PDX tumors, the fore-mentioned cycling hypoxia increases tumor heterogeneity and diversifies growth rates, generating epithelial tumors with rare cell types that may not match the initially grated tumors of the patients [51]. Histological interconversion between LUAD and LUSC or conversion from NSCLC to SCLC has been reported when lung cancer patients are treated with EGFR-tyrosine kinase inhibitor or chemotherapy, but the similarities and differences with our pathologically deviant PDX tumors require further investigation [52-56].
Hypoxia in subcutaneous tumors affect the presence of Epstein- Barr Virus (EBV), which is ubiquitous in the human body [57]. Latent EBVs in lymphocytes that are infiltrated in patient tumors are lytically reactivated by HIF-1a that is activated by tumor hypoxia, which is formed when grafted into immune-deficient NSG mice [58,59]. Human lymphocytes transformed by P1 viral oncogenes can proliferate under normoxic as well as hypoxic conditions, forming XALD [60,61]. As such, tumor hypoxia seems to play a key role in the generation of pathologically unmatched subcutaneous tumors, such as phenotypic conversion of cell types or XALD.
In preclinical, co-clinical, or collateral studies using PDX models as a drug testbed, TME is a key factor determining the responses [62,63]. In PDX models, many murine stromal cells are not functional and B-cells and T-cells do not mature, and no natural killer cells exist, and myeloid cells, such as macrophages and dendritic cells, are defective, leaving murine fibroblasts as the only functional stromal cells in TME [64,65]. Human CAF in the patient tumor fragments is known to be diluted and gradually substituted by murine CAFs during serial passages in NSG mice [66,67]. Therefore, to utilize PDX tumors as a copycat of the patient tumor, not only somatic mutations but also changes in gene expression caused by TME modification need to be analyzed in-depth between patient and PDX tumors.
Unlike patient tumors in which the angiogenic factors are produced and released by tumor cells, CAF and tumor infiltrating immune cells, including lymphocytes and macrophages, in PDX tumors, the main sources of angiogenic factors, are the human tumor cells and the murine CAFs, which require effective cell-cell communication across species. In patient tumors, growing cancer cells are in continuous contact with human fibroblasts from early stages for extended periods of time to form conditioned CAFs, but in PDX tumors, direct abrupt contacts with unconditioned murine fibroblasts may cause prolonged chronic hypoxia and autophagy at an early stage. As a consequence, compared with patient tumors, HIF-1a activation levels were lowered (Table S2) and the weakened angiogenesis may delay tumor growth and decrease tumor engraftment rates, even leading to the failure of tumor formation (Table 2a). In PDX LUSC and LCNEC tumors, some of the angiogenesis-related genes were upregulated, but not in those of LUAD. Maybe this is why LUAD had significantly lower engraftment rates in NSG mice than other cell types (Table 5).
When PDX tumors were used to generate a humanized PDX model, the humanized mouse infused with CD34+ human hematopoietic stem cells have fully differentiated functional human myeloid cells that substitute for the defective dendritic cells and macrophages to partly rescue TME in PDX tumors. Even so, the murine fibroblasts still have to play a role in mediating the crosstalk between the tumor mass and the tumor infiltrating immune cells as well as the endothelial cells [68,69]. Thus, murine fibroblasts make a critical contribution to angiogenesis for the successful growth of grafted human tumors in humanized PDX models as well as in PDX models.
Concordant with a number of previous studies, the pathologically relevant PDX models in this study showed that various somatic mutations, including driver mutations and intra-tumoral heterogeneities, were retained through serial passages, and relative to ANT, the patient and PDX tumors showed similar expression patterns (Table 3, Table 4) [70]. Expanded after serial passages, PDX models have potential for developing targeted drugs in preclinical studies, or in co-clinical or collateral studies for immunotherapies after being grafted to the humanized NSG mice.
Nonetheless, depending on the cell types of the patient tumors, some PDX tumors showed additional somatic mutations, and different fragments of the same patient tumors often showed a variety of growth patterns, which indicated that intratumor heterogeneity might be an inevitable factor causing variation in each individual, even within the specific PDX model. Unlike somatic mutations, differences in gene expression were considerable between the PDX and patient tumors. Therefore, caution is warranted when using PDX models to evaluate the systemic effects of targeted drugs and to translate the outcome into clinical applications.
There have been a number of studies pointing out the intratumor heterogeneity of PDX models [71-74]. To enhance the engraftment efficiencies of PDX tumors, and to minimize the clonal evolution during serial passages, mouse fibroblasts need to be conditioned quickly by the human tumors, with angiogenesis proceeding without delay, and EBV-activation needs to be interrupted effectively [75,76].
Conclusions
Current protocols for PDX model preparation often result in significantly diverse engraftment rates, depending on tumor types, and rare patient tumors that are valuable for clinical research are frequently lost during the process. For PDX models to qualify for wide use in preclinical studies for drug development as well as in collateral studies for clinical benefit, the engraftment rate needs to be elevated, and TME, especially fibroblasts, close to that of the patient tumors needs to be established in the PDX models. Murine fibroblasts, defective murine macrophages, and EBV-infected lymphocytes are some of the key factors contributing to the inefficiency of PDX tumor generation when associated with tumor hypoxia, which may provide basic knowledge for a strategic improvement in graft efficiencies in the future. The presence of pathologically irrelevant subcutaneous tumors, including phenotypic conversions and XALD, was attributed to tumor hypoxia and EBV activation.
Acknowledgements
Professional English writing service was provided by Enago for the manuscript.
Author’s contributions
J Lee wrote most of the manuscript. CH Seo and MY Park performed the genomic analysis. BK Kim assisted the data collection, image preparation and drafting. S-H Kim assisted the manuscript writing and editing. JH Lee, HK Kim, JH Cho, YS Choi, S Shin, J Kim provided the patients’ tumor tissues and clinical information. Y-A Choi prepared the human tumor tissue for xenograft. JH Kang collected patients’ clinical information and performed the H&E and IHC staining.HK Song, and HY Jang prepared the PDX models. J-E Lee supervised the entire process of PDX model development. S Lee supervised the genomic analysis. M Cho and D-S Son performed the statistical analysis of the patients’ clinical information and their association with engraftment efficiency of PDX. J Lee, CH Seo, and BK Kim equally contributed to this study. J Lee, J-E Lee, and J Kim supervised the entire study. All authors read and approved the final manuscript.
Funding
The majority of this work was supported by the Technology Innovation Program of the Ministry of Trade, Industry, and Energy, Republic of Korea (Grant No. 10050154), and partly by DNA Link, Inc.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All patients provided written informed consents authorizing the collection and use of their body tissues for study purposes. This study was approved by the IRB of Samsung Medical Cancer (2014-10-069, 2015-04-018, 2018-03-110).
Consent for publication
All patients provided written informed consents authorizing the collection and use of their body tissues for study purpose.
Competing interest
The authors declare that they have no competing interests.
References
- Barta JA, Powell CA, Wisnivesky JP (2019) Global epidemiology of lung cancer. Ann Glob Health 85(1) [Crossref]
- Park JY, Jang SH (2016) Epidemiology of lung cancer in Korea: recent trends. Tuberc Respir Dis 79(2):58-69.
- Casal-Mouriño A, Ruano-Ravina A, Lorenzo-González M, Rodríguez-Martínez Á, Giraldo-Osorio A, et al. (2021) Epidemiology of stage III lung cancer: frequency, diagnostic characteristics, and survival. Transl Lung Cancer Res 10(1):506.
- Griffin R, Ramirez RA (2017) Molecular targets in non–small cell lung cancer. Ochsner J 17(4):388-392.
- Heist RS, Sequist LV, Engelman JA (2012) Genetic changes in squamous cell lung cancer: a review. J Thorac Oncol 7(5):924-33.
- Ferrara MG, Stefani A, Simbolo M, Pilotto S, Martini M, et al. (2021) Large cell neuro-endocrine carcinoma of the lung: current treatment options and potential future opportunities. Front Oncol 11:650293.
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, et al. (2012) Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9(6):338-350.
- Gengenbacher N, Singhal M, Augustin HG (2017) Preclinical mouse solid tumour models: status quo, challenges and perspectives. Nat Rev Cancer 17(12):751-765.
- Morgan KM, Riedlinger GM, Rosenfeld J, Ganesan S, Pine SR (2017) Patient-derived xenograft models of non-small cell lung cancer and their potential utility in personalized medicine. Front Oncol 7:2. [Crossref]
- Jiang Y, Zhao J, Zhang Y, Li K, Li T, et al. (2018) Establishment of lung cancer patient-derived xenograft models and primary cell lines for lung cancer study. Journal of translational medicine. 16:1-8.
- Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, et al. (2014) Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 4(9):998-1013.
- Fiebig HH, Schuchhardt C, Henss H, Fiedler L, Löhr GW (1984) Comparison of tumor response in nude mice and in the patients. Behring Inst Mitt. 74:343-352.
- Boedigheimer MJ, Freeman DJ, Kiaei P, Damore MA, Radinsky R (2013) Gene expression profiles can predict panitumumab monotherapy responsiveness in human tumor xenograft models. Neoplasia 15(2):125-132.
- Meraz IM, Majidi M, Meng F, Shao R, Ha MJ, et al. (2019) An improved patient-derived xenograft humanized mouse model for evaluation of lung cancer immune responses. Cancer Immunol ResAug 7(8):1267-179.
- Jung HY, Kim TH, Lee JE, Kim HK, Cho JH, et al. (2020) PDX models of human lung squamous cell carcinoma: Consideration of factors in preclinical and co-clinical applications. J Transl Med Dec;18:1-3. [Crossref]
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, DeFelice M, et al. (2012) High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2(1):82-93.
- Weber CM, Henikoff S (2014) Histone variants: dynamic punctuation in transcription. Genes Dev 28(7):672-682.
- Nyström A, Velati D, Mittapalli VR, Fritsch A, Kern JS, et al. (2013) Collagen VII plays a dual role in wound healing. J Clin Invest Aug 123(8):3498-3509.
- Liu Z, Lebrin F, Maring JA, van den Driesche S, van der Brink S, et al. (2014) ENDOGLIN is dispensable for vasculogenesis, but required for vascular endothelial growth factor-induced angiogenesis. PLoS One. 9(1):e86273.
- Horikawa S, Ishii Y, Hamashima T, Yamamoto S, Mori H, et al. (2015) PDGFRα plays a crucial role in connective tissue remodeling. Sci Re. Dec 5(1):17948.
- Zhang J, Cao R, Zhang Y, Jia T, Cao Y, et al. (2009) Differential roles of PDGFR‐α and PDGFR‐β in angiogenesis and vessel stability. FASEB J (1):153-163.
- Coma S, Allard-Ratick M, Akino T, van Meeteren LA, Mammoto A, et al. (2013) GATA2 and Lmo2 control angiogenesis and lymphangiogenesis via direct transcriptional regulation of neuropilin-2. Angiogenesis. 16:939-952. [Crossref][Google Scholar][Pubmed]
- Olszewska-Pazdrak B, Hein TW, Olszewska P, Carney DH (2009) Chronic hypoxia attenuates VEGF signaling and angiogenic responses by downregulation of KDR in human endothelial cells. Am J Physiol Cell Physiol 296(5):C1162-1170.
- Wicks EE, Semenza GL (2022) Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest 132(11)
- Sueyoshi K, Komura D, Katoh H, Yamamoto A, Onoyama T, et al. (2021) Multi-tumor analysis of cancer-stroma interactomes of patient-derived xenografts unveils the unique homeostatic process in renal cell carcinomas. iScience 24(11).
- Folaron M, Merzianu M, Duvvuri U, Ferris RL, Seshadri M (2019) Profiling the stromal and vascular heterogeneity in patient-derived xenograft models of head and neck cancer: Impact on therapeutic response. Cancers. 11(7):951.
- Lugano R, Ramachandran M, Dimberg A (2020) Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci 77:1745-1770.
- El Hout M, Cosialls E, Mehrpour M, Hamai A (2020) Crosstalk between autophagy and metabolic regulation of cancer stem cells. Molecular Cancer 19(1):1-7
- Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F (2019) Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death & Differentiation. 26(4):690-702.
- Wang P, Wan WW, Xiong SL, Feng H, Wu N (2017) Cancer stem-like cells can be induced through dedifferentiation under hypoxic conditions in glioma, hepatoma and lung cancer. Cell Death Discov 3(1):1-10.
- Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, et al (2012) Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci U S A 109(8):2784-9.
- Hassan Venkatesh G, Bravo P, Shaaban Moustafa Elsayed W, Amirtharaj F, Wojtas B, et al ( 2020) Hypoxia increases mutational load of breast cancer cells through frameshift mutations. Oncoimmunology 9(1):1750750.
- Kondo A, Safaei R, Mishima M, Niedner H, Lin X, et al (2001) Hypoxia-induced enrichment and mutagenesis of cells that have lost DNA mismatch repair. Cancer Res 61(20):7603-7.
- Sethi N, Kikuchi O, McFarland J, Zhang Y, Chung M, et al (2019) Mutant p53 induces a hypoxia transcriptional program in gastric and esophageal adenocarcinoma. JCI insight 4(15).
- Bhandari V, Li CH, Bristow RG, Boutros PC (2020) Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat Commun 11(1):737.
- Ando Y, Ta HP, Yen DP, Lee SS, Raola S, et al (2017) A microdevice platform recapitulating hypoxic tumor microenvironments. Sci Rep 7(1):15233.
- Kim I, Choi S, Yoo S, Lee M, Kim IS (2022) Cancer-associated fibroblasts in the hypoxic tumor microenvironment. Cancers 14(14):3321.
- Loh JJ, Ma S (2021) The role of cancer-associated fibroblast as a dynamic player in mediating cancer stemness in the tumor microenvironment. Front Cell Dev Biol 9:727640.
- Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I (2018) The hypoxic tumour microenvironment. Oncogenesis 7(1):10.
- Semenza GL (2017) Hypoxia‐inducible factors: coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J 36(3):252-9.
- López de Andrés J, Grinan-Lison C, Jimenez G, Marchal JA (2020) Cancer stem cell secretome in the tumor microenvironment: a key point for an effective personalized cancer treatment. J Hematol Oncol 13:136.
- Prager BC, Xie Q, Bao S, Rich JN (2019) Cancer stem cells: The architects of the tumor ecosystem. Cell Stem Cell 24:41–53.
- Kugeratski FG, Atkinson SJ, Neilson LJ, Lilla S, Knight JRP, et al. (2019) Hypoxic cancer–associated fibroblasts increase NCBP2-AS2/HIAR to promote endothelial sprouting through enhanced VEGF signaling. Sci Signal 12:eaan8247.
- Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, et al. (2014) VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res 74:1566-1575.
- Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, et al. (2010) HIF1-alpha functions as a tumor promoter in cancer-associated fibroblasts, and as a tumor suppressor in breast cancer cells. Cell Cycle 9:3534-3551.
- Bader SB, Dewhirst MW, Hammond EM (2021) Cyclic hypoxia: An update on its characteristics, methods to measure it and biological implications in cancer. Cancers (Basel) 13:23.
- Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, et al. (2005) Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem 280:41928-41939.
- Pressley M, Gallaher JA, Brown JS, Tomaszewski MR, Borad P, et al. (2021) Cycling hypoxia selects for constitutive HIF stabilization. Sci Rep 11:5777.
- Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, et al. (2019) Molecular landmarks of tumor hypoxia across cancer types. Nat Genet 51:308-318.
- Qiu G-Z, Jin M-Z, Dai J-X, Sun W, Feng J-H, et al. (2017) Reprogramming of the tumor in the hypoxic niche: The emerging concept and associated therapeutic strategies. Trends Pharmacol Sci 38:669-686.
- Peille AL, Vuaroqueaux V, Wong SS, Ting J, Klingner K, et al. (2020) Evaluation of molecular subtypes and clonal selection during establishment of patient-derived tumor xenografts from gastric adenocarcinoma. Commun Biol 3(1):367.
- Xi YZ, Xie L, Tan XW, Zeng SL (2022) Transformation of adenocarcinoma to squamous cell carcinoma as a source of EGFR-TKI resistance: A case report and literature review. Front Oncol 12:942084.
- Jukna A, Montanari G, Mengoli MC, Cavazza A, Covi M, et al.( 2016) Squamous cell carcinoma “transformation” concurrent with secondary T790M mutation in resistant EGFR-mutated adenocarcinomas. Journal of Thoracic Oncology. 11(4):e49-51.
- Kanazawa H, Ebina M, Ino-oka N, Shimizukawa M, Takahashi T, et al. (2000) Transition from squamous cell carcinoma to adenocarcinoma in adenosquamous carcinoma of the lung. Am J Pathol 156(4):1289-1298.
- Sehgal K, Varkaris A, Viray H, VanderLaan PA, Rangachari D, et al. (2020) Small cell transformation of non-small cell lung cancer on immune checkpoint inhibitors: uncommon or under-recognized?. J Immunother Cancer 8(1).
- Oser MG, Niederst MJ, Sequist LV, Engelman JA (2015) Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol 16(4):e165-172.
- Cho EY, Kim KH, Kim WS, Yoo KH, Koo HH, et al. (2008) The spectrum of Epstein-Barr virus-associated lymphoproliferative disease in Korea: incidence of disease entities by age groups. J Korean Med Sc Apr 23(2):185-192.
- Murata T, Sugimoto A, Inagaki T, Yanagi Y, Watanabe T, et al. (2021) Molecular basis of epstein–barr virus latency establishment and lytic reactivation. Viruses. 13(12):2344.
- Kraus RJ, Yu X, Cordes BL, Sathiamoorthi S, Iempridee T, et al. (2017) Hypoxia-inducible factor-1α plays roles in Epstein-Barr virus’s natural life cycle and tumorigenesis by inducing lytic infection through direct binding to the immediate-early BZLF1 gene promoter. PLoS pathogens 13(6):e1006404.
- Price AM, Dai J, Bazot Q, Patel L, Nikitin PA, et al. (2017) Epstein-Barr virus ensures B cell survival by uniquely modulating apoptosis at early and late times after infection. Elife 6:e22509.
- Darekar S, Georgiou K, Yurchenko M, Yenamandra SP, Chachami G, et al. Epstein-Barr virus immortalization of human B-cells leads to stabilization of hypoxia-induced factor 1 alpha, congruent with the Warburg effect.
- Mhaidly R, Mechta‐Grigoriou F (2021) Role of cancer‐associated fibroblast subpopulations in immune infiltration, as a new means of treatment in cancer. Immunol Rev 302(1):259-272.
- Mao X, Xu J, Wang W, Liang C, Hua J, et al. (2021) Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol cancer 20(1):1-30.
- Cao X, Shores EW, Hu-Li J, Anver MR, Kelsail BL, et al. (1995) Defective lymphoid development in mice lacking expression of the common cytokine receptor γ chain. Immunity 2(3):223-238.
- Global epidemiology of lung cancer.
- Yoshida GJ (2020) Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol 13(1):1-6.
- Blomme A, Van Simaeys G, Doumont G, Costanza B, Bellier J, et al. (2018) Murine stroma adopts a human-like metabolic phenotype in the PDX model of colorectal cancer and liver metastases. Oncogene 37(9):1237-1250.
- Linxweiler J, Hajili T, Körbel C, Berchem C, Zeuschner P, et al. (2020) Cancer-associated fibroblasts stimulate primary tumor growth and metastatic spread in an orthotopic prostate cancer xenograft model. Sci Rep 10(1):12575.
- Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, et al. (2020) A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 20(3):174-186.
- Sun H, Cao S, Mashl RJ, Mo CK, Zaccaria S, et al. (2021) Comprehensive characterization of 536 patient-derived xenograft models prioritizes candidates for targeted treatment. Nat Commun 12(1):5086.
- Kanaki Z, Voutsina A, Markou A, Pateras IS, Potaris K, et al. (2021) Generation of non-small cell lung cancer patient-derived xenografts to study intratumor heterogeneity. Cancers 13(10):2446.
- Shi J, Li Y, Jia R, Fan X (2020) The fidelity of cancer cells in PDX models: Characteristics, mechanism and clinical significance. (2020) Int J Cancer 146(8):2078-2088.
- Sprouffske K, Kerr G, Li C, Prahallad A, Rebmann R, et al. (2020) Genetic heterogeneity and clonal evolution during metastasis in breast cancer patient-derived tumor xenograft models. Comput Struct Biotechnol J 18:323-331.
[Crossref][Google Scholar][Pubmed]
- Wellbrock C (2015) Spatial intra‐tumour heterogeneity in acquired resistance to targeted therapy complicates the use of PDX models for co‐clinical cancer studies. EMBO Mol Med 7(9):1087-1089.
- Leiting JL, Hernandez MC, Yang L, Bergquist JR, Ivanics T, et al. (2019) Rituximab decreases lymphoproliferative tumor formation in hepatopancreaticobiliary and gastrointestinal cancer patient-derived xenografts. Sci Rep 9(1):5901.
- Butler KA, Hou X, Becker MA, Zanfagnin V, Enderica-Gonzalez S, et al. (2017) Prevention of human lymphoproliferative tumor formation in ovarian cancer patient-derived xenografts. Neoplasia 19(8):628-636.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi