Review Article, J Clin Genom Vol: 2 Issue: 1
Emerging Techniques in Single-Cell Epigenomics and their Applications to Cancer Research
Lo PK and Zhou Q*
Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, USA
*Corresponding Author : Qun Zhou
Associate Professor, Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, USA
Tel: +1-410-706-1615
Fax: +1-410-706-8297
E-mail: qzhou@som.umaryland.edu
Citation: Lo PK, Zhou Q (2020) Emerging Techniques in Single-Cell Epigenomics and their Applications to Cancer Research. J Clin Genom. 2:1.
Abstract
Epigenomics encompasses studies of the chemical modifications of genomic DNA and associated histones, interactions between genomic DNA sequences and proteins, the dynamics of the chromosomal conformation, the functional relationships between these epigenetic events, and the regulatory impacts of these epigenetic events on gene expression in cells. In comparison to current techniques that are only capable of characterizing average epigenomics features across bulk cell ensembles, singlecell epigenomics methodologies are emerging as powerful new techniques to study cellular plasticity and heterogeneity, as seen in stem cells and cancer. Here we summarize available techniques for studies of single-cell epigenomics, review their current applications to cancer research, and discuss future possibilities. This review also highlights that the full potential of single-cell epigenetic studies will be comprehended through integrating the multi-omics information of genomics, epigenomics and transcriptomics.
Keywords: Epigenomics; Genomic DNA; Oncogenesis; Chemotherapeutic treatments ; single-cell techniques; intratumor heterogeneity
Introduction
Epigenetics covers the study of molecular mechanisms driving heritable changes in gene expression without the involvement of changes in DNA sequences. These epigenetic events involve chemical modifications to DNA and the associated histone proteins, and changes in DNA accessibility and chromatin conformation. In addition to genetic alterations, epigenetic aberrations are another key pathological mechanism driving carcinogenesis. Epigenetic changes in cancer cells alter chromosomal conformation and accessibility to the transcriptional machinery and regulators, which can in turn activate oncogenesis and silence tumor-suppressive mechanisms by modulating the expression of protein-coding and non-coding (e.g. microRNA and long non-coding RNA) genes. These sequential epigenetic alterations lead to uncontrolled growth, invasion and metastasis of cancer cells. In contrast to genetic mutations that are stable and irreversible, epigenetic alterations are dynamic and reversible, leading to epigenetic heterogeneity within tumors during their development or in their responses to environmental stimuli or chemotherapeutic treatments [1]. This epigenomics variability leads to heterogeneity in cancer by creating cell-to-cell differences in the patterning of DNA methylation, histone modifications, or expression of protein coding genes or noncoding RNAs. In spite of the functional relevance of this epigenomics variability in cancer heterogeneity, this variation at the single-cell level is difficult to assess in the past due to a lack of methods capable of analyzing it.
Understanding of epigenetic events in the past was mostly based on studies involving techniques that measure average epigenetic states of bulk cell populations and tissues. Although these studies of bulk cell populations have revealed generalized epigenetic mechanisms (e.g. the roles of epigenetic marks in active or repressed transcriptional states) and established the mapping of epigenetic makeup in many bulk normal and cancerous cell types as well as tissues, it is difficult to use these average profiling measurements to examine epigenetic events occurring in poorly characterized cell populations and rare cell types (e.g. niches) within normal or tumor tissues. Due to the limitations of bulk epigenetic profiles, the study of epigenomics at a single-cell level is necessary. Owing to this need, new techniques for studying single-cell epigenomics have been developed [2,3]. Technological advances in single-cell epigenomics overall involve the development of indexing systems in genome-wide sequencing of the high throughput cell platform and techniques for improving the recovery yields of epigenetic materials isolated from a single cell or from the limited cell number [2]. This article summarizes these new single-cell techniques that measure the modifications of DNA and histones, chromatin accessibility and chromosome conformation, reviews their current applications to cancer research and discusses the future potential of single-cell epigenomics in addressing issues important for understanding of cancer.
Methodologies of Single-Cell Epigenomics
Newly developed single-cell techniques are concisely summarized in this section to provide readers with the overview of advances in single-cell epigenomics. The timeline for the evolution of technologies on single-cell epigenomics is shown in Figure 1. Advantages and disadvantages of these established single-cell epigenomics methods and their comparisons are summarized in Table 1. Comprehensive details of these techniques have been well reviewed elsewhere [2,3].
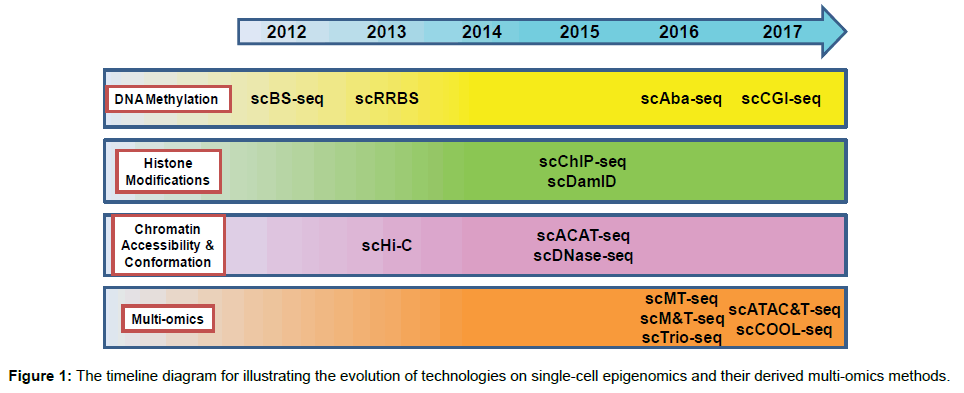
Figure 1: The timeline diagram for illustrating the evolution of technologies on single-cell epigenomics and their derived multi-omics methods.
Suitable for high-throughput The microfluidics method:
The high coverage of sequencing reads
High sequencing resolutionThe combinatorial indexing method:
The low coverage of sequencing reads
Low sequencing resolution The microfluidics method:
Low throughput scDNase-seqHigh sequencing resolutionlow mapping efficiency and lower throughput scNOMe-seq/
scCOOL-seqSimultaneously detect chromatin accessibility and DNA methylationNo selectivity for open chromatin
The lower coverage of genome wide cytosinesChromatin conformationscHi-CA powerful method for defining the individual chromosome organization, compartmentalization, and interchromosomal interactionsLimited resolution
Table 1: Advantages and disadvantages of current developed single-cell epigenomics technologies.
Analysis of single-cell DNA modifications
Methylation at cytosine residues of CpG dinucleotide in the genome is a major type of DNA modifications, which is involved in silencing gene expression. Among methods used to analyze DNA methylation, bisulfite sequencing (BS-seq) that involves the chemical conversion of unmethylated cytosine into uracil is a powerful tool to measure CpG methylation in a single-base resolution [4,5]. Recent technical advances enable BS-seq to measure DNA methylation at a single-cell level. To reach the single-cell level, single cells are isolated and transferred into separate PCR tubes/wells (e.g. the 96- well platform) by serial dilution, micromanipulation (e.g. mouth pipetting), fluorescence-activated cell sorting (FACS), laser-capture microdissection (LCM) and microfluidic devices [6]. The extracted DNA derived from single cells can be processed either by reducedrepresentation bisulfite sequencing (RRBS) or by post-bisulfite adaptor tagging (PBAT) to create indexed sequencing libraries wherein well-specific barcodes that can be traced back to particular single cells are incorporated into bisulfite-converted sequence reads (Figure 2) [7-10]. In the scRRBS method, five DNA processing steps before PCR amplification, including genomic DNA purification, restriction enzyme digestion, end repair/dA tailing, adapter ligation and bisulfite conversion, are prepared in a single tube to avoid loss of single-cell DNA (Figure 2) [7]. To increase the sequencing coverage of methylated DNA, bisulfite treatment in PBAT-based single-cell BS-seq (scBS-seq) is performed before incorporation of indexed sequencing primers into DNA libraries (Figure 2) [8-10]. This new procedure of PBAT prevents the degradation of indexed sequencing libraries caused by bisulfite treatment [8-10]. In addition to these bisulfite-dependent methods, a bisulfite-independent method called genome wide CpG island (CGI) methylation sequencing for single cells (scCGI-seq) has been invented to profile DNA methylation patterns in a single cell, a new technique combining methylationsensitive restriction enzyme digestion and multiple displacement amplification (MDA) for selective detection of methylated CGIs (Figure 2) [11]. The MDA is an experimental step that selectively amplifies methylated CGI-containing long DNA strands but not short unmethylated CGI fragments. These single-cell methods preserve long-range DNA methylation profiles across a sparse, diploid genomic landscape. By comparing scBS-seq to scRRBS, scBS-seq provides the higher coverage of CpG dinucleotide (~3.7 million CpGs) than that of scRRBS (~one million CpGs) [7,9,12]. Therefore, scBS-seq includes more CpG-sparse regions than scRRBS. Nevertheless, the coverage of CGI from scRRBS analysis is higher than that from scBS-seq analysis [7,12]. Moreover, scRRBS profiles the genome in a relatively consistent and less random manner when compared to scBS-seq. This advantage allows scRRBS to provide the higher overlap between the individual CpG sites covered in different single cells [12]. Due to both methods offering their common and unique DNA methylation information, these two techniques are complementary to each other. Therefore, the choice of which method to use relies on the interest of specific studies. With regard to the most recent scCGI-seq, it offers the higher coverage of CGIs (>70% of all CGIs) than that of scRRBS [11]. This improved coverage from scCGI-seq represents a 66-fold increase in the fraction of consistently profiled CGIs across individual cells when compared to scRRBS [11]. Owing to this substantial improvement, scCGI-seq has become a powerful and reliable method to profile DNA methylation of CGIs in a single cell.
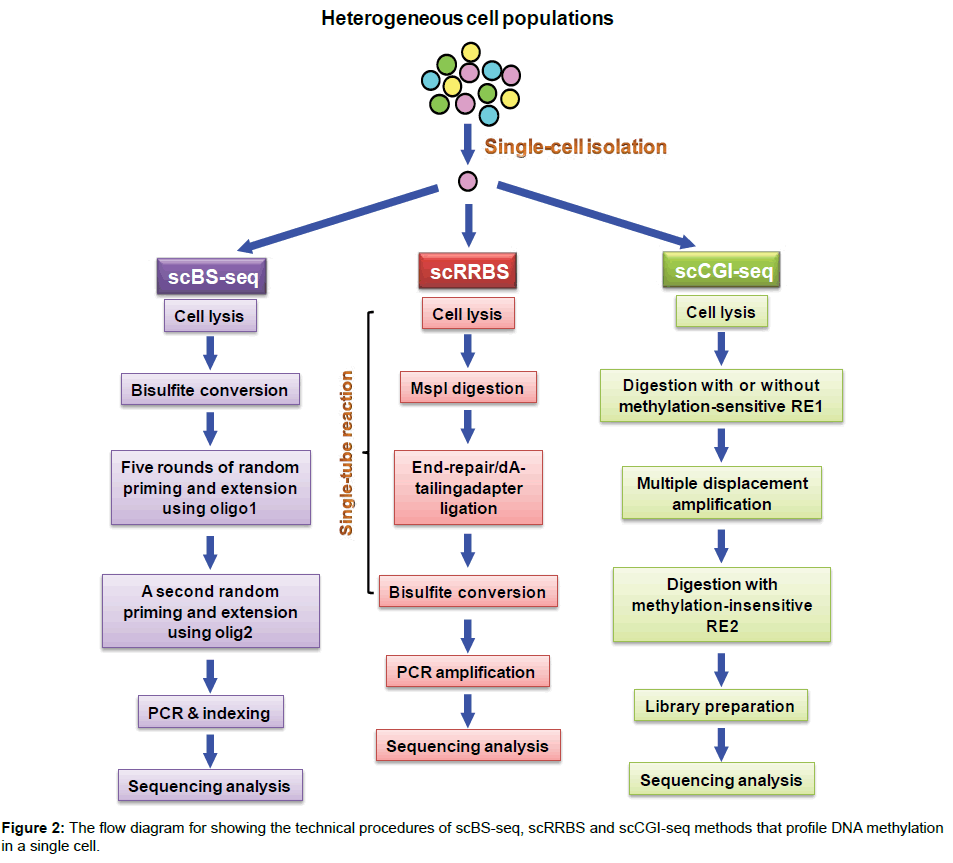
Figure 2: The flow diagram for showing the technical procedures of scBS-seq, scRRBS and scCGI-seq methods that profile DNA methylation in a single cell.
Given that the outcome of DNA methylation is to alter gene expression, concurrent profiling analysis of both DNA methylome and RNA transcriptome at a single-cell level is critical for understanding the relationship between DNA methylation patterns and RNA expression profiles in a single cell. Recently, two techniques, called scMT-seq and scM&T-seq, have been established for parallel single-cell sequencing analysis of both methylome and transcriptome [13,14]. scMT-seq is a combination of both scRRBS and scRNAseq (Smart2-seq) and scM&T-seq is a technique integrating scBS-seq with scRNA-seq [13,14]. Both single-cell techniques have taken the advantage of the method capable of simultaneously isolating cytosolic RNA for RNA-seq and nuclear genomic DNA for DNA methylome profiling [13,14]. These multi-omics methods have opened a window of opportunity for dissecting the mechanisms of epigenetic gene regulation.
In addition to methylated cytosine (5mC), the other minor types of DNA modifications have been identified, such as hydroxymethylated cytosine (5hmC) that is generated by oxidation of 5-methylcytosine, a reaction mediated by ten-eleven translocation (TET) methylcytosine dioxygenase. 5hmC has been known to contribute to the active demethylation of the paternal chromosomes during early embryonic stages [15]. The currently established methods for measuring these minor DNA modifications, such as TET-assisted bisulfite sequencing (TAB-seq) and Aba-seq, could potentially be compatible with singlecell approaches. Indeed, Mooijman and colleagues presented a single-cell Aba-seq technology, based on 5hmC glucosylation and glucosylation-dependent digestion of DNA, to analyze genome-wide and strand-specific 5hmC. Their studies have revealed chromosomewide cell-to-cell variability in 5hmC profiles [16-18].
Analysis of single-cell histone modifications
The various covalent modifications of histones have been discovered and they are functionally involved in regulating genomic characteristics and transcriptional states [19]. These histone marks are routinely mapped using chromatin immunoprecipitation sequencing (ChIP-seq), which involves chromatin immunoprecipitation with antibodies specific to these histone marks. Although this method is powerful, high background noise is problematic for performing ChIP-seq at the single-cell level. To overcome this issue, genomic DNA from a pool of single cells is processed through micrococcal nuclease (MNase) digestion and barcoding steps before the immunoprecipitation step. This improved approach enables pulldown to be effectively performed on thousands of cells. Although the low coverage of sequencing reads is the disadvantage of scChIP-seq, this limited coverage potentially enables this technique to concurrently assess a large number of single cells. Indeed, by combination with a droplet-based microfluidics setup, this approach can process large numbers of single cells in parallel [20].
In addition to single-cell ChIP-seq (scChIP-seq), another method called DamID has been developed to map protein-DNA interactions at a single-cell level. This new method involves the expression of a fusion protein of Escherichia coli deoxyadenosine methylase (Dam) and the interested protein in cells. This design allows Dam to methylate DNA on adenine residues in close proximity to the sites of protein binding. Methylated sites are then digested by the methylation-sensitive restriction enzyme DpnI, followed by ligation with sequencing adapters to generate sequencing libraries. This new method has been successfully applied to the study of genomic DNA interactions with the nuclear lamina in single cells [21]. Moreover, single-cell DamID could be an alternative for genome-wide analysis of histone modifications by fusing Dam with specific histone readers or modifiers. Due to the current sequencing resolution of singlecell DamID limited to the order of 100 kb, this disadvantage to some extent restricts its application. Therefore, future optimizations and improvements are needed for enabling this technique to map transcription factor binding sites in single cells.
Analysis of single-cell chromatin accessibility and conformation
Transcriptional activation is known to result in disruption of nucleosome organization at promoters, enhancers, silencers, insulators and locus control regions due to transcription factor binding. Therefore, these regulatory DNA regions coincide with open/accessible genomic sites of remodeled chromatin. Mapping of these accessible chromatin regions is relevant to understanding of regulation of gene expression, cell proliferation, cell differentiation, functional diversification and disease development. Several techniques have been developed to map accessible chromatin regions in a single cell. The first mapping method called the assay for transposase-accessible chromatin using sequencing (ATACseq) is based on the employment of a Tn5 transposase enzyme to simultaneously cleave DNA within accessible open chromatin regions and the followed ligation of fragmented DNA with adapter sequences (a process called tagmentation) [22,23]. To achieve the single-cell level, ATAC-seq adopts a “combinatorial indexing” strategy in which the tagmentation is performed on 96 pools of a few thousand nuclei with the incorporation of a unique barcode into each pool and then these 96 pools are mixed and split again before a second barcode is incorporated into each new pool via polymerase chain reaction (PCR) (Figure 3). Optimized two-round barcode indexing enables a particular barcode combination to be specific to a single cell (Figure 3) [24]. Therefore, this approach allows the ATAC-seq data to represent the epigenomics information of a single cell. Another modified singlecell ATAC-seq method employs a microfluidics device to generate single-cell pools instead of using the combinatorial indexing method [25]. Although its throughput is substantially lower, this modified technique has substantially improved the sequencing resolution and coverage, encompassing 70,000 reads per cell compared with 3000 reads from the prior method.
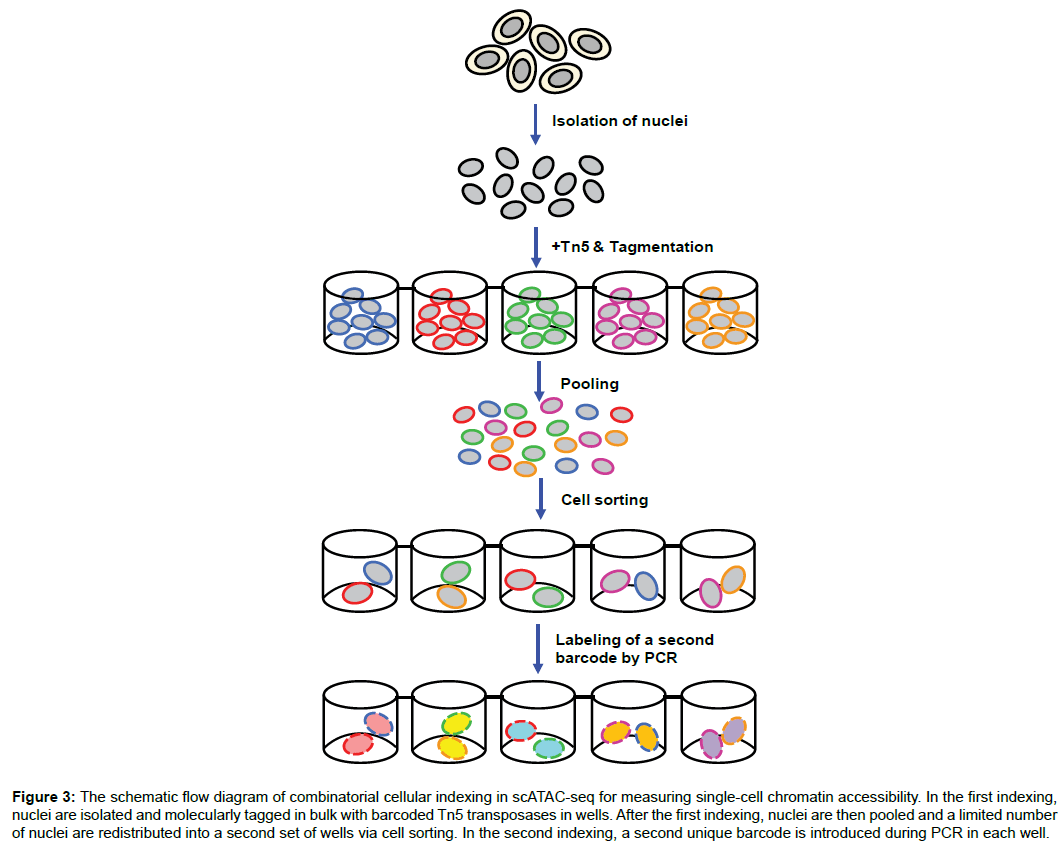
Figure 3: The schematic flow diagram of combinatorial cellular indexing in scATAC-seq for measuring single-cell chromatin accessibility. In the first indexing, nuclei are isolated and molecularly tagged in bulk with barcoded Tn5 transposases in wells. After the first indexing, nuclei are then pooled and a limited number of nuclei are redistributed into a second set of wells via cell sorting. In the second indexing, a second unique barcode is introduced during PCR in each well.
In addition to the transposase-based method, a DNase I-based single-cell sequencing (scDNase-seq) has been applied to analysis of chromatin accessibility in a single cell [26]. The design of this method is based on the fact that open/accessible chromatin regions are vulnerable to DNase I digestion. This technique offers a high sequencing resolution (300,000 mapped reads per single cell), but low mapping efficiency and lower throughput are its disadvantages. In particular, scDNase-seq and aforementioned scATAC-seq could be potentially conducted in parallel with scRNA-seq through the effective separation of DNA from RNA or parallel amplification of both materials [14,27]. Moreover, an attractive technique called nucleosome occupancy and methylome sequencing (NOMe-seq) is capable of simultaneously analyzing nucleosome positioning, accessible chromatin and DNA methylation on a genome-wide scale [28]. NOMe-seq exploits a methyltransferase enzyme to methylate GpC dinucleotides within open/accessible genomic regions, but not those within nucleosome-bound DNA regions. After the GpC methylation reaction, DNA is treated with bisulfite and processed as BS-seq for next-generation sequencing analysis. Due to the difference between enzymatic (GpC) and endogenous (CpG) DNA methylation, NOMe-seq is able to map both methylation patterns in a single-base resolution. By integrating scNOMe-seq with PBAT-based scBS-seq, a novel multi-omics method called single-cell COOL-seq has been developed for parallel analysis of the chromatin state/nucleosome positioning, DNA methylation, copy number variation and ploidy in single cells (Figure 4) [29]. scCOOL-seq has been utilized to analyze the reprogramming of the chromatin state and DNA methylation in mouse preimplantation embryos [29]. This new multi-omics methodology offers the first single-cell and parental allele-specific analysis of the global chromatin state and DNA methylation dynamics at single-base resolution in early mouse embryos and provides new insights into this embryonic process involving the heterogeneous yet highly ordered features of epigenomics reprogramming.
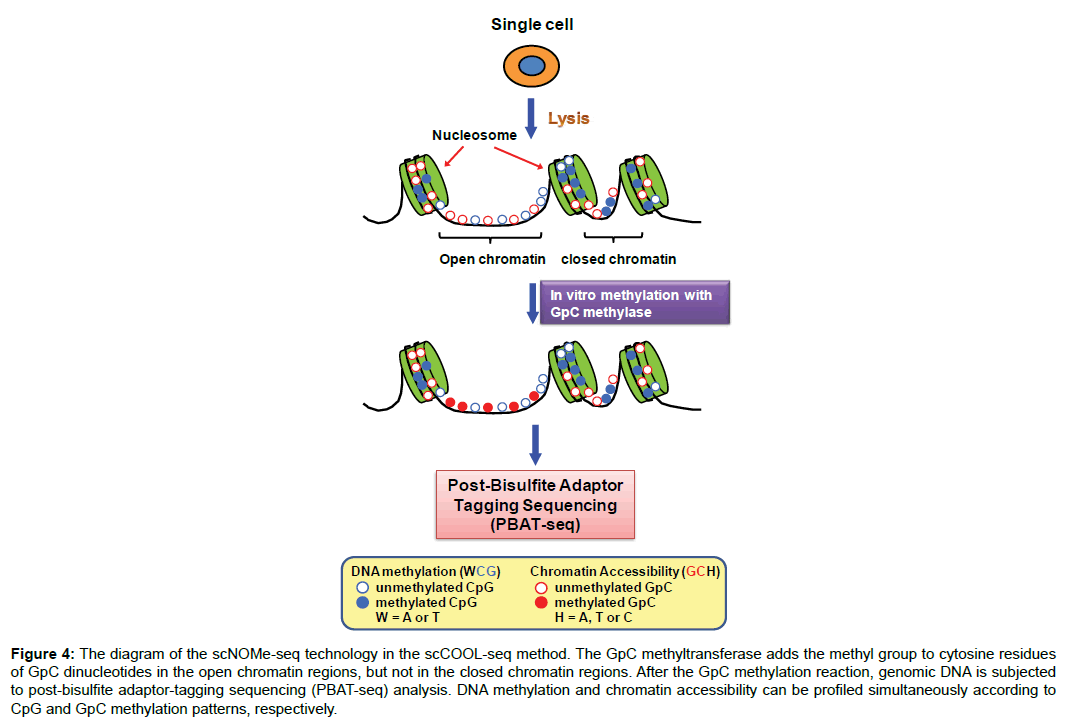
Figure 4: The diagram of the scNOMe-seq technology in the scCOOL-seq method. The GpC methyltransferase adds the methyl group to cytosine residues of GpC dinucleotides in the open chromatin regions, but not in the closed chromatin regions. After the GpC methylation reaction, genomic DNA is subjected to post-bisulfite adaptor-tagging sequencing (PBAT-seq) analysis. DNA methylation and chromatin accessibility can be profiled simultaneously according to CpG and GpC methylation patterns, respectively.
Due to no selectivity for open chromatin, the high depth of sequencing may be necessary for scNOMe-seq to guarantee coverage of genomic regions of interest. Moreover, given the need of filtering out some ambiguous nucleotide positions (e.g. C-C-G and G-C-G), this reduces the coverage of genome-wide cytosines from scNOMeseq analysis by ~50% when compared to that from scBS-seq analysis. However, despite this reduced coverage, scNOME-seq is still able to profile a large proportion of the genomic loci with important regulatory roles (e.g. promoters and enhancers). In addition to characterizing linear chromatin organization, studies of chromosomal conformation have become important as the three-dimensional topology of chromosomes determines the interactions of gene promoters with enhancers, silencers, and insulators. A genome-scale technique called Hi-C has been developed to define the conformation of chromosomes. In Hi-C, cellular chromatin DNA is fixed and then digested by sequence-specific restriction enzymes while maintaining intact protein–protein and protein-DNA interactions. Re-ligation of digested DNA ends generates DNA chimaeras, which preserve spatial proximity interactions between different genomic loci. Given that Hi-C analysis of bulk cell populations may result in some ambiguity in interpretation of the results, single-cell Hi-C (scHiC) has been developed as a cutting-edge technique to assess chromosomal conformation in a single cell [30,31]. scHi-C is currently limited in its resolution but still able to define the individual chromosome organization, compartmentalization, and interchromosomal interactions.
Single-cell epigenomics and cancer
The study of intratumor heterogeneity is important as it relates to tumor microenvironment, genetic as well as epigenetic diversity resulting from clonal evolution of tumor cells, and cancer progression to invasive/metastatic disease (Figure 5) [32]. To determine whether intratumor heterogeneity can be defined by their genomic, epigenomic and transcriptomic profiles, single-cell sequencing technologies are mandatory. Indeed, single-cell DNA sequencing (scGenome-seq) was successfully employed to reveal that multiple cancer types can undergo clonal evolution, and to identify founder mutations and subclonal mutations that are implicated in cancer progression (Figure 5) [33,34]. Similarly, single-cell transcriptome profiling has been exploited to discover stem-cell-like subpopulations within cancers (Figure 5) [35,36]. These single-cell-based studies have provided new insights into how cancer progresses and whether single-cell profiling can be used to predict disease outcome.
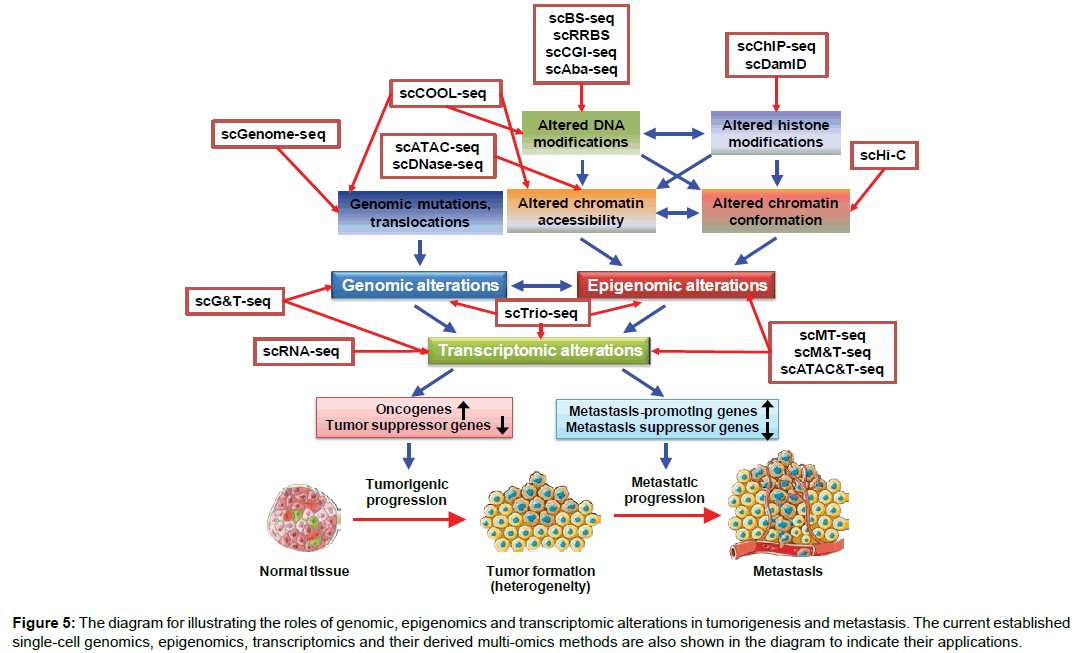
Figure 5: The diagram for illustrating the roles of genomic, epigenomics and transcriptomic alterations in tumorigenesis and metastasis. The current established single-cell genomics, epigenomics, transcriptomics and their derived multi-omics methods are also shown in the diagram to indicate their applications.
The cancer epigenome has been known to be substantially remodeled during tumorigenesis, metastasis and drug resistance (Figure 5). As therapeutic drugs targeting epigenetic enzymes (e.g. DNA methyltransferases and histone deacetylases) have been demonstrated to be effective in treatment of several cancer types, these indicate that epigenomic alterations play a critical role in disease progression [37]. It has been shown that DNA hypomethylation occurs on a global scale in cancer. In contrast, DNA hypermethylation occurs at specific genomic loci, which are associated with aberrant nucleosome positioning and chromatin modifications. Recently, genome-wide profiling of methylome in triple-negative breast cancers (TNBCs) has stratified TNBCs into three distinct methylation clusters, which are associated with patients’ prognosis [38]. Nevertheless, the important question regarding whether intratumor heterogeneity can be reflected by epigenetic heterogeneity can only be addressed by single-cell epigenomic analyses. Two recent studies have reported the potential of single-cell epigenomic techniques in addressing the role of epigenetic heterogeneity in cancer. By employing a singlecell triple omics sequencing technique called scTrio-seq that can simultaneously assess the genomic copy-number variations (CNVs), DNA methylome, and transcriptome of 25 single cancer cells, Hou and colleagues have identified two subpopulations present within cells isolated from a human hepatocellular carcinoma tissue sample [39]. Another study by Litzenburger and colleagues employed scATAC-seq combined with scRNA-seq to reveal that in K562 leukemic cells, the levels of the cell surface marker CD24 correlated with chromatin accessibility changes linked to GATA transcription factors in single cells [40]. Their finding is clinically relevant as the status of GATA/CD24 in leukemic cells was related to drug sensitivity and differential self-renewal capacity. Their study suggests that GATA factors are critically involved in determining cell phenotypes through modulating epigenomic plasticity. Evidence from these single-cell epigenomic studies demonstrates that single-cell epigenomic features can be utilized to characterize intratumor heterogeneity and its roles in drug sensitivity and the clonal evolution of cancer cells.
Perspectives
Numerous single-cell epigenomic technologies have been developed to profile the distinct types of epigenomic alterations occurring at a single-cell level in heterogeneous tumor tissue (Figure 5). By combining these single-cell epigenomic techniques with other omics technologies, several multi-omics methods (e.g. scMT-seq, scM&T-seq, scATAC&T-seq, scCOOL-seq, scTrioseq) have been established to analyze two to three different omics in a same single cell (Figure 5). It can be imagined that new multiomics technologies with ability to analyze genomic, epigenomic and transcriptomic profiles in parallel will be rapidly developed in the future to identify rare subpopulations of cancer cells within a tumor. Some of aforementioned studies have demonstrated this possibility. Studies of cancer stem cells (CSCs) are clinically important as CSCs have been considered to contribute to intratumor heterogeneity, invasive progression, metastasis, and drug resistance. Due to dynamic self-renewal, differentiation and dormancy of CSCs in the tumor microenvironment, single-cell epigenomic techniques will be powerful tools to effectively trace CSC plasticity in a tumor. Singlecell technologies will be also very useful to analyze multi-omic profiles of circulating tumor cells (CTCs), representing metastatic cancer cells, in comparison to those from primary tumors. Multi-omic analyses of CTCs are crucial for understanding the mechanisms underlying cancer metastasis. Moreover, single-cell epigenomic studies may potentially guide the development of novel therapeutics targeting subpopulations with clinical significance in cancer. Particularly, single-cell analysis of DNA methylation is an attractive strategy for cancer screening as the DNA methylation information in the genome is more stable than RNA that is used in transcriptomic profiling.
References
- Friedmann-Morvinski D, Verma IM (2014) Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep 15: 244-253.
- Schwartzman O, Tanay A (2015) Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet 16: 716-726.
- Kelsey G, Stegle O, Reik W (2017) Single-cell epigenomics: Recording the past and predicting the future. Science 358: 69-75.
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215-219.
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315-322.
- Yaron JR, Ziegler CP, Tran TH, Glenn HL, Meldrum DR (2014) A convenient, optimized pipeline for isolation, fluorescence microscopy and molecular analysis of live single cells. Biol Proced Online 16: 9.
- Guo H, Zhu P, Wu X, Li X, Wen L, et al. (2013) Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 23: 2126-2135.
- Miura F, Enomoto Y, Dairiki R, Ito T (2012) Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res 40: e136.
- Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, et al. (2014) Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods 11: 817-820.
- Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schönegger A, et al. (2015) Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep 10: 1386-1397.
- Han L, Wu HJ, Zhu H, Kim KY, Marjani SL, et al. (2017) Bisulfite-independent analysis of CpG island methylation enables genome-scale stratification of single cells. Nucleic Acids Res 45: e77.
- Guo H, Zhu P, Guo F, Li X, Wu X, et al. (2015) Profiling DNA methylome landscapes of mammalian cells with single-cell reduced-representation bisulfite sequencing. Nat Protoc 10: 645-659.
- Hu Y, Huang K, An Q, Du G, Hu G, et al. (2016) Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol 17: 88.
- Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, et al. (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13: 229-232.
- Peat JR, Dean W, Clark SJ, Krueger F, Smallwood SA, et al. (2014) Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep 9: 1990-2000.
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, et al. (2012) Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368-1380.
- Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, Wang H, Sun D, Cheng X,, Zhu Z PS, Zheng Y (2013) High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep 3: 567-576.
- Mooijman D, Dey SS, Boisset JC, Crosetto N, van Oudenaarden A (2016) Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol 34: 852-856.
- Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7-18.
- Rotem A, Ram O, Shoresh N, Sperling R, Goren A, et al. (2015) Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 33: 1165-1172.
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, et al. (2015) Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134-147.
- Adey A, Morrison HG, Asan, Xun X, Kitzman JO, et al. (2010) Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol 11: R119.
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213-1218.
- Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, et al. (2015) Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348: 910-914.
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, et al. (2015) Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486-490.
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, et al. (2015) Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528: 142-146.
- Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A (2015) Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol 33: 285-289.
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, et al. (2012) Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 22: 2497-2506.
- Guo F, Li L, Li J, Wu X, Hu B, et al. (2017) Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res 27: 967-988.
- Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, et al. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502: 59-64.
- Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, et al. (2015) Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc 10: 1986-2003.
- Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, et al. (2015) Toward understanding and exploiting tumor heterogeneity. Nat Med 21: 846-853.
- Navin NE (2015) The first five years of single-cell cancer genomics and beyond. Genome Res 25: 1499-1507.
- Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, et al. (2014) EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov 4: 956-971.
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, et al. (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396-1401.
- Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, et al. (2015) Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 526: 131-135.
- Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11: 726-734.
- Stirzaker C, Zotenko E, Song JZ, Qu W, Nair SS, et al. (2015) Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun 6: 5899.
- Hou Y, Guo H, Cao C, Li X, Hu B, et al. (2016) Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26: 304-319.
- Litzenburger UM, Buenrostro JD, Wu B, Shen Y, Sheffield NC, et al. (2017) Single-cell epigenomic variability reveals functional cancer heterogeneity. Genome Biol 18: 15.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi