Research Article, J Proteomics Enzymol Vol: 6 Issue: 2
Evaluation of Tryptic Peptides from Neisseria meningitidis Outer Membrane Proteins PorA and PorB Digestion, Peptides
Gail Whiting*, Clive Metcalfe, Adrian F Bristow and Jun X Wheeler
National Institute for Biological Standards and Control, Medicine and Healthcare Products Regulatory Agency, Blanche Lane, South Mimms, Potters Bar, Hertfordshire EN6 3QG, United Kingdom
*Corresponding Author : Gail Whiting
National Institute for Biological Standards and Control (NIBSC) Blanche Lane, South Mimms, Potters Bar, EN6 3QG, UK
Tel: 01707641203
E-mail: gail.whiting@nibsc.org
Received: July 14, 2017 Accepted: August 04, 2017 Published: August 06, 2017
Citation: Whiting G, Metcalfe C, Bristow AF, Wheeler JX (2017) Evaluation of Tryptic Peptides from Neisseria meningitidis Outer Membrane Proteins PorA and PorB. J Proteomics Enzymol 6:2. doi: 10.4172/2470-1289.1000133
Abstract
Abstract
Peptide-based protein quantitation using LC-MS/MS is a valuable method for the measurement of specific proteins in complex biological samples. To ensure accurate measurement of the target protein the signature peptides must be true stoichiometric representatives of the amount of protein in the sample. We report here the results of our evaluation of candidate signature peptides for IDMS quantitation of Neisseria meningitidis outer membrane proteins PorA and PorB. We observed considerable differences in the quantitation of the individual peptides and determined that the digestion protocol used was crucial to the accuracy of protein quantitation. A rapid rate of cleavage was essential to produce high quality data for the quantitation of PorA and PorB. This report also highlights the need for signature peptides and their isotopically labelled form, i.e. the internal standards, to be stable throughout digestion. However, we did observe that biased quantitation likely due to peptide instability could be mitigated by concurrent addition of the labelled internal standard peptide. Furthermore we observed that sample matrices have differing effects on specific peptide cleavage during protein digestion. In conclusion, the selection of stable signature peptides and the optimisation of the digestion conditions are not trivial considerations but are essential for accurate protein quantitation.
Keywords: Isotope dilution mass spectrometry; Signature peptide; Protein quantitation; Trypsin digestion
Introduction
The development of targeted mass spectrometry approaches including multiple reactions monitoring (MRM) have improved the quantitation of given target proteins in a complex biological matrix [1-3]. For MRM-based quantitation, specific “signature” peptides are considered to be stoichiometric representatives of the protein from which they are cleaved, and thus it follows that one mole of measured peptide will equate to one mole of intact protein. The use of stable isotopically labelled, heavy peptides as internal standards (IS) combined with MRM provides the highest level of confidence in measurement precision [4,5]. IS are synthesised as labelled analogues by replacing 12C14N with 13C15N isotopes to increase the protein or peptide mass without changing the chemical properties. The drawback of isotope dilution mass spectrometry (IDMS) is that any bias in the final yield of the signature peptides will have an enormous impact on the accuracy of quantitation. Studies have shown that, depending on the conditions for digestion, the quantitation of different signature peptides from the same protein may be variable [6,7] and that this can lead to uncertainty in the measurement. Peptide instability also introduces error as the amount of measured peptide is underrepresented. As stability is unique to each peptide it is essential to consider the properties of each tryptic peptide when selecting candidates for quantitation [7,8]. Post-translational modification introduces variability in the mass of a peptide therefore peptides containing N-linked glycosylation consensus sequences and peptides containing methionine and tryptophan which can be oxidised should be avoided [9,10]. Trypsin is the most commonly used proteolytic enzyme used in mass spectrometry-based proteomics. The cleavage efficiency of trypsin has a dramatic effect on the release of tryptic peptides through non-specific and missed cleavages [11,12]. This can be minimised by optimising incubation conditions and trypsin concentration. Furthermore, peptides containing ArgPro and LysPro sequences are resistant to cleavage by trypsin and should be avoided. Different sample matrices have peptide, time and matrix specific effects on protein digestion [13]. The matrix can influence reproducibility of sample preparation and can affect the performance of the trypsin digestion on peptide formation, for example between individual human serum samples [11,14]. Various strategies to eliminate matrix effect have been applied with varying degrees of success, e.g. spin filter devices or immunoaffinity capture [15-17]. Despite the potential interference of the matrix on peptides, in solution digests provide the highest recovery and lowest quantitation variation with the least peptide generation bias [18]. MRM has the potential to quantify the amount of antigen in vaccines derived from outer membrane vesicle (OMV) preparations. Neisseria meningitidis serogroup B (MenB) OMVs contain a large number of proteins which represent a challenging and complex milieu for MS-based quantitation [19]. In this study we evaluate the conditions required to generate signature peptides for the accurate quantitation of PorA, the major antigenic protein, and PorB, the most abundant outer membrane protein found in MenB OMVs.
Materials and Methods
Screening for PorA and PorB signature peptides
N. meningitidis serogroup B strain NZ98/254 PorA (NMB1429) and PorB (NMB2039) protein sequences were downloaded from www.uniprot.org and tryptic peptides with 7 or more and 20 or less residues were identified in-silico using Pinpoint version 1.2 (Thermo Scientific, Waltham, MS, USA). Redundant peptides and peptides occurring more than once in the database were eliminated by filtering against Neisseria meningitidis serogroup B serotype 15, strain H4476. As a result, 13 PorA and 12 PorB peptides were evaluated by using tryptic digests of purified PorA protein or OMV preparations to determine those most suitable for MRM-MS. Pinpoint version 1.2 was used to generate target m/z lists with automatic prediction of parent ion (Q1) and product ion (Q3) m/z transitions and collision energies using defined charge states (precursor charge state 2 and product charge state 1) and y-ion fragmentation type. Between four and five transitions for each peptide were selected manually by excluding product ion transitions of low and high masses (<300 and <1000). The initial MRM method, containing the complete list of target peptides, transitions and collision energies, was exported in csv format as a non-scheduled method so that all the transitions were scanned in the triple quadrupole mass spectrometer (TSQ Quantum, Thermo Scientific) throughout the entire chromatography run. The retention times for each of the peptides were established from the data acquired from the non-scheduled method. In the next iteration, using MRM, the transitions for each of the peptides of interest in a 2 min retention time window were monitored to identify peptides with good signal intensities for parent and fragment ion transitions. Four iterations were performed by taking the raw data file from the previous round and selecting the transitions with the best signal.
Synthesis of native and labelled peptides
PorA and PorB paired peptides, consisting of a native and a heavy (labelled) analogue, were custom synthesized by Thermo Fisher Scientific. Ten nmol (±5%) of each peptide, determined by AAA, was supplied at 5 pmol/μL in 5% (v/v) acetonitrile. The heavy analogues of the PorA and PorB target peptides were made by incorporating a 13C and 15N labelled amino acid at a position close to the carboxyterminal of the peptide. These are shown in Table 1. Synthetic peptides were stored at -70°C and once defrosted were not refrozen and were discarded after one month.
| Signature peptide | Peptide sequence | Precursor ion (m/z) | ms/ms ion 1 | ms/ms ion 2 | ms/ms ion 3 | Collision energy (eV) |
|---|---|---|---|---|---|---|
| PorA_P1N | ISDFGSFIGFK | 609.3 | 902.5 | 755.4 | 464.3 | 24 |
| PorA_P1L | ISDFGSFIGFK | 612.8 | 909.5 | 762.4 | 471.3 | 24 |
| PorA_P2N | GSEDLGEGLK | 502.7 | 731.4 | 616.4 | 503.3 | 20 |
| PorA_P2L | GSEDLGEGLK | 506.3 | 738.4 | 623.4 | 510.3 | 21 |
| PorA_P3N | SAYTPAHVVVNK | 700.4 | 672.4 | 573.3 | 474.3 | 27 |
| PorA_P3L | SAYTPAHVVVNK | 703.4 | 678.4 | 579.3 | 480.3 | 27 |
| PorA_P4N | FGNAVPR | 380.7 | 613.3 | 556.3 | 442.3 | 16 |
| PorA_P4L | FGNAVPR | 383.7 | 619.4 | 562.3 | 448.3 | 16 |
| PorA_P5N | TSAIVSGAWLK | 566.8 | 760.4 | 661.4 | 574.3 | 23 |
| PorA_P5L | TSAIVSGAWLK | 570.3 | 767.5 | 668.4 | 581.4 | 23 |
| PorB_P1N | GQEDLGNGLK | 515.8 | 845.4 | 716.4 | 488.3 | 21 |
| PorB_P1L | GQEDLGNGLK | 519.3 | 852.5 | 723.4 | 495.3 | 21 |
| PorB_P2N | SDYLGVNK | 448.2 | 693.4 | 530.3 | 417.2 | 19 |
| PorB_P2L | SDYLGVNK | 451.2 | 699.4 | 536.3 | 423.3 | 19 |
| PorB_P3N | FVSTAGGVGLR | 532.3 | 817.5 | 629.4 | 558.3 | 21 |
| PorB_P3L | FVSTAGGVGLR | 535.8 | 824.5 | 636.4 | 565.4 | 22 |
Table 1: Signature peptides from PorA and PorB target proteins.
Preparation of native and labelled calibration standards
The native peptide calibration standard was prepared by combining the native PorA and PorB peptides to a final concentration of 0.5 pmol/μL in 0.1% formic acid. The internal standard (IS) was prepared in the same way using the heavy PorA and PorB peptides.
Calibration curves for the quantitation of the PorA and PorB peptides were generated using seven standard solutions containing 15, 30, 50, 70, 100, 180 and 250 fmol/μL of the native peptide calibration standard and 70 fmol/μL of the paired heavy peptides as IS.
Evaluation of peptide performance in different matrices
N. meningitidis serogroup B NZ94/254 outer membrane vesicle (OMV) preparations either in solution at approximately 1 mg/mL protein concentration or adsorbed to aluminium hydroxide were provided by the group in Bacteriology, NIBSC. Recombinant PorA from Escherichia coli was a gift from Dr Hannah Chan and Dr Sunil Maharjan, NIBSC. Protein concentration by amino acid analysis was 4 μg/mL (C.A.T. GmbH and Co, Tübingen, Germany). For in-solution digestions 10-40 μL aliquots of OMV or rPorA were diluted with 50 mM ammonium bicarbonate buffer, pH 8.0 (ABC) to 1 to 5 μg of OMV protein (bulk and vaccine product) or 0.1 to 0.5 μg rPorA. In some experiments ABC was substituted with 200 mM triethylammonium bicarbonate (TEAB). Denaturation was performed by adding RapiGest surfactant (Waters, Bedford, MA, USA), solubilised in buffer and heating at 100°C for up to 10 min. The samples were incubated for 2 to 24 h with 100 pmol/digest of sequencing grade modified trypsin (Promega, Madison, WI, USA). As a result trypsin was present in ratio of 1:2 for OMV in solution and 1:5 for OMV adsorbed to aluminium hydroxide. For LysC/ trypsin double digestions, 100 pmol/digest sequencing grade LysC (Promega) was added to the sample with the trypsin. Hydrochloric acid was added to 45 mM to degrade the acid labile surfactant. The heavy peptide mixture was added to a final concentration of 70 pmol/μL either immediately prior to addition of trypsin or immediately prior to addition of 0.1% formic acid to a final volume of 0.1 mL. Samples were vortexed and centrifuged briefly, except for the aluminium adsorbed samples which were centrifuged for 5 min, before being transferred to autosampler vials for MRM-MS analysis. The heavy peptides were added as internal standards to tryptic digests of the vaccine and to the native peptide standards. Quantitation was achieved by comparing the peak area of the heavy peptide with that of the synthetic native peptide to generate a standard curve of the endogenous target peptide generated from proteolytic cleavage of the target protein. Quantitation was performed using Quanbrowser module of the Xcalibur v. 2.2 software (Thermo Scientific). In experiments where the effect of reduction/alkylation was examined, OMV samples were boiled with 0.1% Rapi Gest for 5 min. Tris(2- carboxyethyl)phosphine) (TCEP) was added to a final concentration of 5 mM and samples were heated at 60°C for 30 min. After cooling, iodoacetamide was added to a final concentration of 15 mM and the samples were incubated in the dark at room temperature for 30 min prior to trypsin digestion and MRM-MS analysis as described above. Protein bands were excised from Coomassie Blue stained SDSPAGE gels and subsequently de-stained with repeated incubations with 50% acetonitrile (ACN) in 50 mM ABC. Once the gel band was completely de-stained it was washed sequentially with 50 mM ABC, 50% ACN in 50 mM ABC and 100% ACN. In-gel tryptic digestion was carried out at room temperature overnight using 0.5 μg of trypsin in 50 mM ABC for each gel band. Peptides were extracted sequentially using 1% TFA, 50% ACN in 0.2% TFA and 100% ACN. The pooled extractions were dried in a centrifugal evaporator, resuspended in 0.1% formic acid, and heavy peptide mixture was added to 70 fmol/μL before being transferred to autosampler vials for MRM-MS analysis.
Quantitation of signature peptides by LC-MRM-MS
MRM-MS was performed with a TSQ Quantum Access (Thermo Scientific) equipped with an ESI and source operated in positivemode ESI. A spray voltage of +3500 V was used with a heated ion transfer tube setting of 235°C. MRM experiments were performed with both Q1 and Q3 resolution settings at unit resolution on both mass spectrometers (FWHM = 0.7 Da). The machine was operated in positive ion mode with multiple reaction monitoring (MRM) m/z transitions chosen specifically for the PorA and PorB peptides. Instrument control was performed with Xcalibur software (Thermo Scientific, v. 2.2). Peptide separations were performed on a Dionex Ultimate 3000 system. Ten μL of sample was injected via full loop injection onto a 150 mm × 1 mm i.d. Symmetry 300 reverse phase C18 (3.5 μm particle size, Waters) with column temperature set at 40°C. Separation was achieved with a 60-min gradient and a flow rate of 50 μL/min. For the first five min the mobile phase was 100% buffer A (98% water, 2% acetonitrile and 0.1% formic acid). From 5 to 20 min Buffer B (98% acetonitrile, 2% water and 0.1% formic acid) was increased to 23% and then to 30% by 25 min. At 27 min the gradient was increased to 100% B and held for 7 min and then reverted back to 100% A to equilibrate the column.
Determination of digest efficiency by LC-MS/MS
Nano-LC-MS/MS was performed using a U3000 direct nano system coupled with nano-electrospray and LTQ-Orbitrap Discovery mass spectrometer (Thermo). Digests were separated on a PepMap C18 reversed phase nano column (3 μm, 100Ǻ, 50 cm length, Thermo) under a column flow rate of 0.3 μl/min using a linear gradient of 5 -0 25% for 180 min, 25 – 32% for 20 min and 32 – 90% for 10 min of 95% acetonitrile and 0.1% formic acid. MS scan and MS/MS fragmentation were carried out in Orbitrap and LTQ respectively, using 3 cycles of top 5 data-dependent acquisitions with collision-induced dissociation fragmentation and dynamic exclusion mode enabled and total cycle time set at approximately 30 milliseconds. Proteome Discoverer v. 1.4 (Thermo Scientific) and PEAKS v.7 software (Bioinformatics Solutions Inc, Waterloo, Canada) were used for mass spectra processing and database searching of N.meningitidis strain NZ-05/33, which is of the same clonal origin of NZ98/254. Initial mass tolerances by MS were set to 10 ppm and up to two missed tryptic cleavages were considered. Methionine oxidation was set as dynamic modification whereas carboxymethylation on cysteine was set as a static modification. Peptides at rank 1 with high confidence were considered to be unambiguously sequenced.
Results
Generation of PorA and PorB signature peptide standard curves
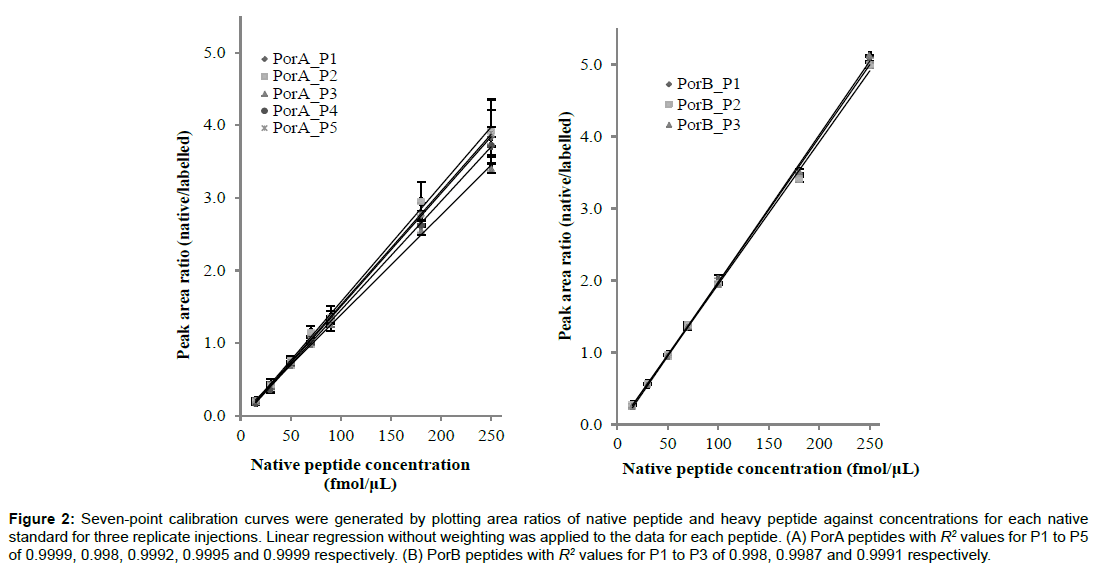
The amino acid sequences of PorA and PorB and the signature peptides selected for use in MRM-MS on the basis of high trypsin cleavage probability, lack of amino acids susceptible to post translational modification (PTM) and good signal intensities of their parent and fragment ion transitions are shown in Figure 1. For PorA, five peptides were selected for synthesis as native and labelled species: ISDFGSFIGFK (PorA_P1), GSEDLGEGLK (PorA_ P2), SAYTPAHVVVNNK (PorA_P3), FGNAVPR (PorA_P4) and TSAIVSGAWLK (PorA_P5). PorA_P1 and PorA_P2 are immediately adjacent to one another; the remaining peptides are well separated across the PorA amino acid sequence. Three peptides that are distally separated on the amino acid sequence of PorB were selected for synthesis: GQEDLGNGLK (PorB_P1), SDYLGVNK (PorB_P2) and FVSTAGGVGLR (PorB_P3). PorA_P5 was retained in the evaluation process, despite containing a potentially variable tryptophan because it is well separated from the other peptides in the PorA sequence. The amounts of both the native and aqua variants of the peptides were accurately determined by AAA. Standard curves constructed by plotting the linear regression analysis response factor (area native peptide/area heavy peptide) versus native peptide concentration are shown in Figure 2. The linear response extends over the entire standard range for each of the peptides with R2 values greater than 0.990.
Figure 2: Seven-point calibration curves were generated by plotting area ratios of native peptide and heavy peptide against concentrations for each native standard for three replicate injections. Linear regression without weighting was applied to the data for each peptide. (A) PorA peptides with R2 values for P1 to P5 of 0.9999, 0.998, 0.9992, 0.9995 and 0.9999 respectively. (B) PorB peptides with R2 values for P1 to P3 of 0.998, 0.9987 and 0.9991 respectively.
Trypsin digest incubation times
Signature peptide performance
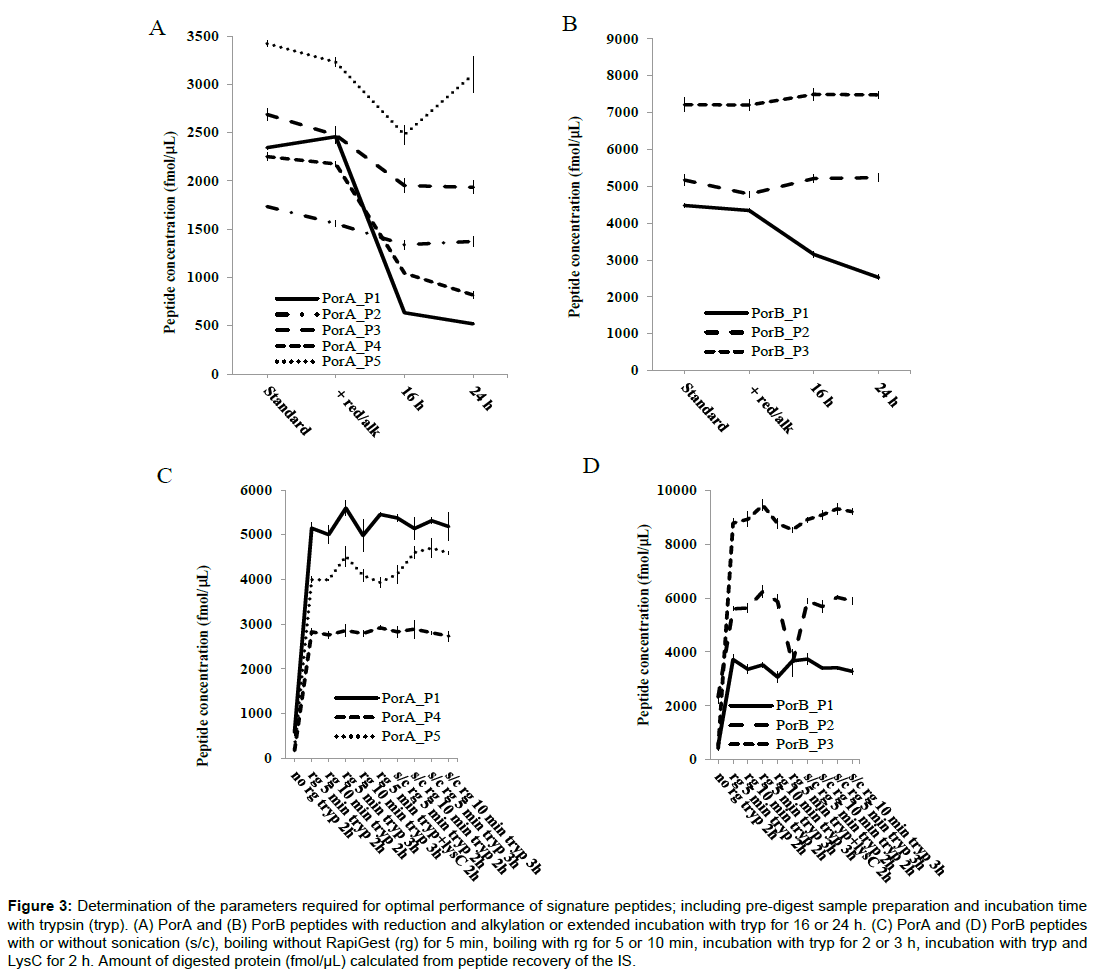
The performance of the digest over different incubation periods was examined by comparing the amount of each of the PorA and PorB signature peptides liberated by the digest (Figure 3). An increase in the incubation period from 2 to 3 h produced no change in the amount of each of the peptides quantified. However, when the incubation period was extended to 16 and 24 h the recovery of some of the peptides was markedly affected. Recovery of PorA peptides PorA_P1 and PorA_P4 were severely diminished, whilst PorA_P3 was less severely affected. For PorB peptides only PorB_P1 showed reduced recovery. The results of the short versus extended incubation showed that it was appropriate to use a high concentration of trypsin over a short incubation period. Combining Lys-C with trypsin in a double digest, did not increase recovery of any of the peptides demonstrating that the digestion after 2 h was likely to be complete.
Figure 3: Determination of the parameters required for optimal performance of signature peptides; including pre-digest sample preparation and incubation time with trypsin (tryp). (A) PorA and (B) PorB peptides with reduction and alkylation or extended incubation with tryp for 16 or 24 h. (C) PorA and (D) PorB peptides with or without sonication (s/c), boiling without RapiGest (rg) for 5 min, boiling with rg for 5 or 10 min, incubation with tryp for 2 or 3 h, incubation with tryp and LysC for 2 h. Amount of digested protein (fmol/μL) calculated from peptide recovery of the IS.
Peptide coverage and non-specific cleavages
Trypsin digestion of the OMV proteins was analysed by LCMS/ MS and both peptide coverage and the number of incomplete cleavage products were used determine the completeness of the digestion (Table 2). Comparison of digests incubated for short periods of time (2-3 h) with digests incubated for more extended periods (16-24 h) showed a reduction both in the number of peptides identified and in the protein sequence coverage for PorA and PorB for the extended incubations. The total number of proteins identified in the digests of OMV decreased after extended incubation as did the total number of matched peptides. The number of mis-cleavages for the PorA and PorB proteins was low, regardless of incubation time, indicating that for PorA and PorB at least, the digest efficiency is not affected by the incubation time. The effect of incubation time on the signature peptides was determined by analysing the LC-MS/ MS in PEAKS 8.0 proteomics suite. A 20-fold reduction in PSMs for PorA_P1 and a 2-fold reduction for PorA_P2 and PorA_P3 were observed; the number of PSMs for the remaining peptides was not significantly altered with different incubation times. No PSMs were observed for PorA_P4 for any of the conditions tested. PSMs for nonspecific cleavages were observed for PorA_P3, SAYTPAHVVVNN and SAYTPAHVVVN. No other non-specific or missed cleavages were observed for the signature peptides, despite using excess trypsin.
| Protein or peptide | Digestion protocol | ||||||
|---|---|---|---|---|---|---|---|
| Control: RG 5 min; 2h tryp | RG 10 min; 2h tryp | RG 5 min; 3h tryp | RG 10 min; 3h tryp | RG 5 min; 16h tryp | RG 5 min; 24h tryp | ||
| No. of peptides identified | PorA | 32 ± 1.4 | 37 ± 1.4 | 35 ± 2.8 | 36 ± 0 | 41 ± 2.8 | 51.5 ± 0.7 |
| Peptide coverage (%) | PorA | 78 ± 0 | 82 ± 0 | 80 ± 2.8 | 79 ± 1.4 | 74.5 ± 0.7 | 75.5 ± 2.1 |
| No. of PSMs identified with no. of mis-cleavages shown in brackets | PorA_P1 | 63.5 ± 6.4 | 61.0 ± 0 | 53.5 ± 9.2 | 48.5 ± 2.1 | 2.5 ± 0.7 | 12.5 ± 13.4 |
| (0) | (0) | (0) | (0) | (0) | (0) | ||
| PorA_P2 | 14 ± 1.4 | 13.5 ± 2.1 | 12.5 ± 3.5 | 11.5 ± 0.7 | 6.5 ± 0.7 | 6 ± 4.2 | |
| (3.5 ± 0.7) | (7.0 ± 1.4) | (7.5 ± 2.1) | (5.5 ± 4.9) | (9 ± 4.2) | (8 ± 0) | ||
| PorA_P3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| - | - | - | - | - | - | ||
| PorA_P4 | 35.5 ± 2.1 | 25.5 ± 14.8 | 28 ± 5.7 | 25 ± 5.7 | 31 ± 5.7 | 45.5 ± 19.1 | |
| (0) | (0) | (0) | (0) | (0) | (0) | ||
| PorA_P5 | 0 | 0 | 1.5 ± 2.1 | 1.5 ± 2.1 | 0 | 1 ± 1.4 | |
| - | - | (0) | (0) | - | (0) | ||
| No.of peptides identified | PorB | 42.5 ± 3.5 | 43.5 ± 0.7 | 43.5 ± 0.7 | 45 ± 0 | 51 ± 1.4 | 63 ± 1.4 |
| Peptide coverage (%) | PorB | 71.5 ± 3.5 | 76 ± 2.8 | 72 ± 1.4 | 72.5 ± 6.4 | 68 ± 2.8 | 69.5 ± 2.1 |
| No. of PSMs identified with no. of mis-cleavages shown in brackets | PorB_P1 | 10.5 ± 3.5 | 8.5 ± 0.7 | 10.5 ± 2.1 | 12.5 ± 2.1 | 10.5 ± 2.1 | 4.5 ± 2.1 |
| (0) | (0) | (0) | (0) | (0) | (0) | ||
| PorB_P2 | 2.0 ± 2.8 | 6 ± 0 | 3 ± 2.8 | 4 ± 1.4 | 2 ± 2.8 | 4 ± 2.8 | |
| (0) | (0) | (0) | (0) | (0) | (0) | ||
| PorB_P3 | 64.5 ± 0.7 | 72 ± 4.2 | 71 ± 1.4 | 66.5 ± 4.9 | 69.5 ± 10.6 | 86.5 ± 0.7 | |
| (0) | (0) | (0) | (0) | (0) | (0) | ||
Table 2: Optimisation of trypsin digestion parameters using LC-MS/MS.
Sample preparation conditions
In order to obtain the complete digestion of PorA and PorB in OMV preparations a number of sample preparation steps are required, including sonication, all of which are likely to have an impact on the accuracy of protein quantitation. A summary of the effects of the different conditions investigated is shown in Figure 3. To determine the effect of detergent assisted denaturation on the liberation of PorA and PorB peptides after tryptic digestion we used the acid labile mass spectrometry compatible detergent RapiGest. Digestion of rPorA and OMV samples in the absence of RapiGest led to a significant reduction in the yield of the selected peptides (Figure 3). In the presence of RapiGest, the recovery of each of the individual peptides was unchanged when we doubled the concentration from 0.1 to 0.2%. Increasing the boiling time of samples prior to digestion from 5 to 10 min also had no effect upon peptide recovery. Denaturation of proteins is often improved by reduction and alkylation of any disulphide bonds that are present. The published sequences of NZ98/254 PorA and PorB do not contain cysteine residues that could contribute to disulphide bonding; however we wanted to investigate whether reduction of disulphide bonds in other outer membrane proteins could improve the solubilisation of the OMV. When OMV samples were reduced with TCEP and subsequently alkylated with iodoacetamide prior to trypsin digestion, we found a small, but insignificant, reduction in the quantitation of the majority of PorA and PorB peptides, compared to the untreated OMVs (Figure 3) indicating reduction and alkylation is unnecessary.
Peptide modifications
PTM of the endogenous target peptides would result in a shift in both their mass and retention time. During the selection of the PorA and PorB signature peptides, those that contained sequences known to be associated with N-linked glycosylation were excluded. PorA _P5 has a tryptophan residue that could be susceptible to oxidation, so tryptophan oxidation transitions were generated using Pinpoint and were scanned for over the length of chromatograph. A peak with an 8 Dalton mass shift which eluted at a different retention time was observed and contributed up to 8% of the total peak area for the synthetic peptide. However the peak was not observed in OMV digests, suggesting the synthetic peptides are undergoing oxidation when combined in the calibration standard solutions before use. In order to avoid over-estimation of protein this species has to be qualified and added to the total amount of the target peptide observed. PorA _P1 contains Asp (D) which is susceptible to dehydration to form a cyclic imide intermediate that can cause cleavage of the peptide chain, particularly in acidic conditions. Working stock solutions of synthetic labelled and native peptides are stored in 0.1% FA where they may degrade over time. We observed that the quantitation of PorA_P1 peptide increased after prolonged storage of the peptide standards suggesting the synthetic peptides are degrading under these conditions. This does not occur in endogenous peptides released by the trypsin digest as the incubation time is short. A situation where the reference peptides are degrading and the target peptides are not will lead to increased protein estimation over time.
Peptide stability during digestion
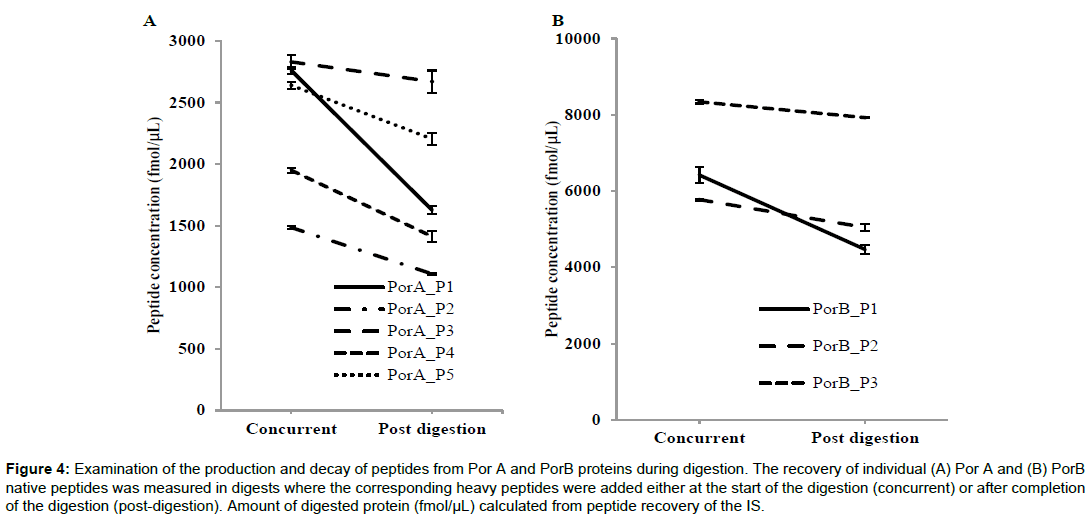
When the quantitation method is applied to the complex OMV samples we noted inconsistencies in quantitation values between the different target peptides for both PorA and PorB (Figure 4). We examined if the time-point when the IS was added had an influence on the quantitation of each peptide, by either adding the heavy peptide mixture at the same time as the trypsin (concurrent) or adding the standard after the digestion (post-digestion). Quantitation of the individual target peptides for PorA and PorB showed a closer correlation when the ISTD was added concurrently both for the bulks and final fills (Figure 4) suggesting that some degradation was occurring during tryptic digestion. The overall quality of the trypsin digest was assessed by LC-MS/MS using total peptide coverage and the number of missed cleavages as markers of digest quality (Table 3). The IS peptides are solubilised in 0.1% formic acid, so the concurrent addition of the IS with trypsin will lower the pH to 2. As trypsin activity is optimal at pH 8 we compared digestions when peptides were adding with trypsin or post-digestion. Substituting ABC with TEAB changes the pH of the digest 2 to 5. For both PorA and PorB, coverage was the same under all the conditions and the number of missed cleavages was low for all indicating the digestion is equally efficient in the pH range of 2 - 5.
Figure 4: Examination of the production and decay of peptides from Por A and PorB proteins during digestion. The recovery of individual (A) Por A and (B) PorB native peptides was measured in digests where the corresponding heavy peptides were added either at the start of the digestion (concurrent) or after completion of the digestion (post-digestion). Amount of digested protein (fmol/μL) calculated from peptide recovery of the IS.
| Concurrent IS ABC (pH = 3) |
Post-digest IS ABC (pH = 8) |
Concurrent IS TEAB (pH = 5) |
Post-digest IS TEAB (pH = 8) |
|
|---|---|---|---|---|
| PorB | ||||
| Peptide coverage (%) | 60.3 ± 0 | 59.6 ± 1.1 | 58.0 ± 2.6 | 60.7 ± 1.1 |
| Total no. of peptides matched | 13 ± 0 | 12.3 ± 0.8 | 12.3 ± 1.3 | 13 ± 0.7 |
| 0 miscleavages | 12 ± 0 | 11.8 ± 0.4 | 11.3 ± 0.8 | 12 ± 0.7 |
| 1 miscleavage | 1 ± 0 | 0.5 ± 0.5 | 1 ± 0.7 | 1 ± 0 |
| 2 miscleavages | 0 | 0 | 0 | 0 |
| PorA | ||||
| Peptide coverage (%) | 69.2 ± 0 | 73 ± 0 | 71 ± 3.2 | 73.3 ± 0.1 |
| Total no. of peptides matched | 17 ± 0 | 17 ± 0 | 18.8 ± 0.8 | 18.3 ± 0.4 |
| 0 miscleavages | 15 ± 0 | 16 ± 0 | 16 ± 0.7 | 16 ± 0.8 |
| 1 miscleavage | 1 ± 0 | 0.25 ± 0.4 | 1.3 ± 0.4 | 0.8 ± 0.8 |
| 2 miscleavages | 1 ± 0 | 1 ± 0 | 2 ± 0.7 | 2 ± 0 |
| OMV digest | ||||
| Total no. of proteins identified | 70 ± 7.5 | 80.5 ± 8.1 | 77.8 ± 8.1 | 78 ± 8.5 |
| No. of peptides matched | 343.8 ± 34.4 | 382 ± 17.6 | 73.3 ± 16.7 | 380 ± 24.4 |
| 0 miscleavages | 323 ± 28.8 | 357.3 ± 15.3 | 341 ± 16.4 | 348.3 ± 22.0 |
| 1 miscleavage | 19.3 ± 6.0 | 22 ± 2.7 | 30.8 ± 2.9 | 30.8 ± 4.6 |
| 2 miscleavages | 1.5 ± 0.5 | 0.3 ± 0.4 | 1.8 ± 0.8 | 1 ± 1.2 |
Table 3: Investigation of effect of concurrent versus post-digestion addition of IS.
Effect of matrix
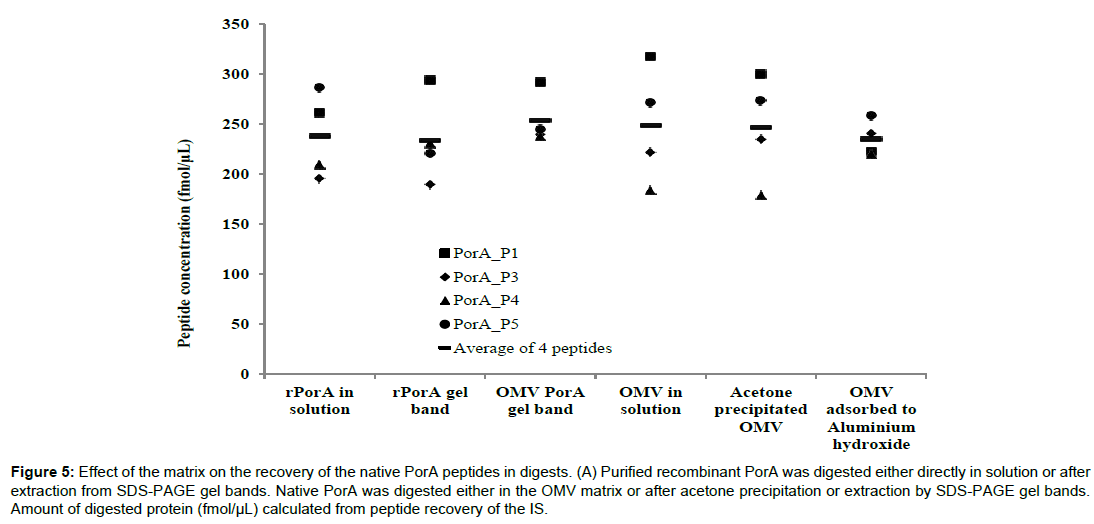
We investigated whether the sample matrix in which the proteins are suspended could have an impact on the recovery of each signature peptide. PorA and PorB proteins were recovered from gel slices, following separation of OMV or recombinant PorA by SDS-PAGE, to act as “matrix-free” samples. We compared the peptide recovery from these “matrix-free” proteins to those from native OMV, acetone-precipitated OMV and OMVs adsorbed onto aluminium (Figure 5). The pattern of recovered peptides for both in-solution and gel band recovered rPorA were very similar, as was the average concentration across the four peptides analysed. However the observed concentrations for PorA peptides analysed from SDS separated OMV showed much less variation compared to in-solution digested OMV (which was similar to the recombinant samples) but again the average concentration of the four peptides was similar. Where the OMV is adsorbed onto alum, less variation in concentration is observed across the four PorA peptides in solution digests compared to the bulk OMV in solution digests.
Figure 5: Effect of the matrix on the recovery of the native PorA peptides in digests. (A) Purified recombinant PorA was digested either directly in solution or after extraction from SDS-PAGE gel bands. Native PorA was digested either in the OMV matrix or after acetone precipitation or extraction by SDS-PAGE gel bands. Amount of digested protein (fmol/μL) calculated from peptide recovery of the IS.
Discussion
Peptide-based protein quantitation using IDMS-MS is a valuable method for the measurement of specific proteins in complex biological samples and therefore this study was initiated to develop a suitable method for the accurate and precise quantitation of the PorA and PorB proteins. However, the selection of stable signature peptides and the optimisation of the digestion conditions are not trivial considerations but are essential for accurate protein quantitation. We report here the results of our evaluation of candidate signature peptides for IDMS quantitation of N. meningitidis outer membrane proteins PorA and PorB. The accuracy of IDMS-MRM to measure concentration of the target protein relies on the signature peptides being true stoichiometric representatives of the amount of protein in the sample. For this to be achieved the target protein must be completely digested with equimolar release of the signature peptides. Several reaction conditions need to be optimised to ensure complete digestion PorA and PorB by trypsin: Solubilisation of the proteins, the substrate to enzyme ratio, the reaction pH and the reaction temperature, the time of reaction and effect of the biological matrix. The complete digestion of PorA and PorB in OMV preparations is challenging, firstly because the hydrophobic nature of membrane proteins can affect solubility and secondly because OMV preparations, either in solution or adsorbed onto aluminium, are subject to aggregation and sample homogeneity can adversely affect peptide quantitation. Results from our study suggest that alum binding may promote better digestion, resulting in improved recovery of the tryptic peptides, due to protein conformational changes [20]. Traditionally the proteomic analysis of hydrophobic proteins has used solubilisation with denaturants such as UREA and SDS prior to digestion. However, removal of these LC-MS/MS incompatible reagents is likely to adversely affect quantitation through sample loss as a result of excess handling. The use of RapiGest, a mass spectrometry compatible detergent [21-23] was previously shown to be essential in achieving consistent and reproducible results for IDMS quantitation of haemagglutinin in influenza virus [24,25]. Similarly for OMVs, the addition of RapiGest facilitates the solubilisation of the sample prior to digestion with trypsin. Tryptic digestion protocols for IDMS deviate significantly from those traditionally used for protein identification as both the endogenous native peptide and the labelled internal standard are required to be stable throughout to prevent quantitation bias through the under or over-estimation of the target protein. For each individual signature peptide the final amount yielded from the trypsin digest is strongly dependent on conditions employed in the digestion. For MRM-based assays of selected peptides this can have an enormous impact on detection limits and quantitation [23]. Significant nonspecific cleavage by trypsin (i.e. not exclusively C-terminal to arginine and Lysine residues) is thought to occur during digestion and this may contribute to inaccurate quantitation as signature peptides may be cleaved. Traditional digestion methods for protein identification purposes typically use enzyme-to-substrate ratios of 1:10 to 1:100 and extended incubation times of 16 to 24 h however for IDMS quantitation excess trypsin (2.5:1) was used in a shortened incubation restricted to 2 h [24]. For OMV preparations adding excess trypsin produced the maximum recovery of the PorA and PorB signature peptides and complete cleavage of the target proteins in their complex biological matrix. The method was also rapid, minimising deaminidation [25] and changes in peptide concentration resulting from sample evaporation, both of which are difficult to quantity and not easily corrected. We identified discrepancies in the quantitation values obtained for the individual PorA and PorB signature peptides that could be due to different rates of production and decay of the signature peptides during the digestion as has been reported previously [7]. Since final quantitation is based on the ratio of target peptide to IS, concurrent addition of the IS should account for the differences in the rates of production and decay as both native and IS would be affected equally in the digest. Our results showed that addition of the heavy peptides concurrently increases the yield of all of the PorA and B signature peptides. This effect is more pronounced with some of the signature peptides and would suggest that for these peptides production during the tryptic digest may happen slowly and at the same time there may be rapid peptide decay during the digestion process. Thus the amount of native peptide at the end of the digest will be less than the amount of peptide originally in the undigested protein. We conclude that compared to the addition of heavy peptides after the digestion, where only those losses of target peptides in subsequent sample injection and chromatography steps would be accounted for, adding the peptides into the digest concurrently gives a more accurate quantitation result. The results that we have presented here highlight the challenges involved in designing a method to quantify multiple proteins within a complex sample as complete digestion of each of the protein targets is essential for accurate quantitation. It is likely that the nature of each individual protein may need to be taken into account when selecting digest conditions. Our finding that trypsin can digest PorA to completeness using conditions traditionally considered to be sub-optimal was not unexpected given that [26-28] previously showed that PorA is surface-exposed and completely accessible to digestion. However, for other proteins, further investigation is required to establish the optimum conditions for complete digestion to be achieved so as not to affect the yield of signature peptides. We can conclude that the results of our analysis of the behaviour of PorA and PorB tryptic peptides in a complex digest show that IDMS is suitable to develop as a method to quantify PorA and PorB.
Acknowledgements
This work was funded by the National Institute for Biological Standards and Control. The authors would like to thank NIBSC colleagues Hannah Chan and Sunil Maharjan for their generous gifts of recombinant PorA and PorB. We are grateful to our colleagues Caroline Vipond, Ian Feavers and Rory Care, and to Stefano Gotta.
References
- Holman SW, Sims PFG, Eyers CE (2012) The use of selected reaction monitoring in quantitative proteomics. Bioanalysis 4: 1763-1786.
- Lange V, Picotti P, Domon B, Aebersold R (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4: 222.
- Picotti P, Aebersold R (2012) Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature Methods 9: 555-566
- Barr JR, Maggio VL, Patterson Jr DG, Cooper GR, Henderson LO, et al. (1996) Isotope dilution-mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem 42: 1676-1682.
- Ong SE, Mann M (2005) Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol 1: 252-262.
- Arsene CG, Ohlendorf R, Burkitt W, Pritchard C, Henrion A, et al. (2008) Protein quantification by isotope dilution mass spectrometry of proteolytic fragments: cleavage rate and accuracy. Anal Chem 80: 4154-4160.
- Shuford CM, Sederoff RR, Chiang VL, Muddiman DC (2012) Peptide production and decay rates affect the quantitative accuracy of protein cleavage isotope dilution mass spectrometry (PC-IDMS). Mol Cell Proteomics 11: 814-823.
- Scott KB, Turko IV, Phinney KW (2015) Quantitative performance of internal standard platforms for absolute protein quantification using multiple reaction monitoring-mass spectrometry. Anal Chem 87: 4429-4435.
- Mo W, Ma Y, TakaoT, Neubert TA (2000) Sequencing of oxidized methionine-containing peptides for protein identification. Rapid Commun Mass Spectrom 14: 2080-2081.
- Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS (2003) Oxidative post-translational modification of tryptophan residues in cardiac mitochondrial proteins. J Biol Chem 278: 19587-19590.
- Burkhart JM, Schumbrutzki C, Wortelkamp S, Sickmann A, Zahedi RP (2012) Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J Proteomics 75: 1454-1462.
- Picotti P, Aebersold, Domon B (2007) The implications of proteolytic background for shotgun proteomics. Mol Cell Proteomics 6: 1589-1598.
- Arnold SL, Stevison F, Isoherranen N (2016) Impact of sample matrix on accuracy of peptide quantification: assessment of calibrator and internal standard selection and method validation. Anal Chem 88: 746-53
- Van de Broek I, Romijin FPHTM, Smit NPM, van der Laarse A, Drifhout JW, et al. (2015) Quantifying protein measurands by peptide measurements: where do errors arise? J Proteome Res 14: 928-942.
- Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, et al. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J Proteome Res 3: 235-244.
- Berna MJ, Zhen Y, Watson DE, Hale JE, Ackermann BL (2007) Strategic use of immunoprecipitation and LC/MS/MS for trace-level protein quantification: myosin light chain 1, a biomarker of cardiac necrosis. Anal Chem 79: 4199-4205.
- Manza LL, Stamer SL, Ham A-JL, Codreanu SG, Liebler DC (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5: 1742-1745.
- Leon IR, Schwämmle V, Jensen ON, Sprenger RR (2013) Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol Cell Proteomics 12: 2992-3005.
- Tani C, Stella M, Donnarumma D, Biagini M, Parente P, et al. (2014) Quantification by LC-MSE of outer membrane vesicle proteins of the Bexsero vaccine. Vaccine 32: 1273-1279.
- Jones LS, Peek LJ, Power J, Markham A, Yazzie B, et al. (2005) Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. J Biol Chem 280: 13406-13414.
- Chen EI, Cociorva D, Norris JL, Yates III JR (2007) Optimization of mass spectrometry compatible surfactants for shotgun proteomics. J Proteome Res 6: 2529-2538.
- Chen EI, McClatchy, Park SK, Yates III JR (2008) Comparisons of mass spectrometry compatible surfactants for global analysis of mammalian brain proteome. Anal Chem 80: 8694-8701.
- Proc JL, Kuzyk MA, Hardie DB, Yang J, Smith DS, et al. (2010) A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J Proteome Res 9: 5422-5437.
- Williams TL, Pirkle JL, Barr JR (2008) Simultaneous quantification of hemagglutinin and neuraminidase of influenza virus using isotope dilution mass spectrometry. Vaccine 26: 2510-2520.
- Williams TL, Luna L, Guo Z, Cox NJ, Pirkle JL, et al. (2012) Quantification of influenza virus hemagglutinins in complex mixtures using isotope dilution tandem mass spectrometry. Vaccine 30: 2475-2482.
- Norrgran J, Williams TL, Woolfitt AR, Solano MI, Pirkle JL, et al. (2009) Optimization of digestion parameters for protein quantification. Anal Chem 393: 48-55.
- Hao P, Ren Y, Alpert AJ, Sze SK (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol Cell Proteomics 10: O111.009381.
- Tsolakos N, Brookes C, Taylor S, Gorringe A, Tang CM, et al. (2014) Identification of vaccine antigens using integrated proteomic analyses of surface immunogens from serogroup B Neisseria meningitidis. J Proteomics 101: 63-76.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi