Research Article, J Virol Antivir Res Vol: 10 Issue: 5
Genome sequencing, Analysis and Characterization of Baculovirus Infecting the Caterpillar, Spilosoma Obliqua Walker (Arctiidae) (Insecta: Lepidoptera) from India
Sayan Paul1,2, Subburathinam Balakrishnan1, Arun Arumugaperumal1, Emmanuel Joshua Jebasingh Sathiya Balasingh Thangapandi1, Huidrom Sarjubala Devi3, Thang Johnson3, Shyam Maisnam3, Sandhya Soman Syamala1, Ramamoorthy Sivakumar4, Raman Karthikeyan4, Chandramohan Subburaman5, Sathyalakshmi Alaguponniah6, Deepa Velayudhan Krishna6, Krishnan Nallaperumal6, Salam Shantikumar Singh7, Muthukumaran Azhaguchamy5, Jeyaprakash Rajendhran4, Varatharajan Ramaiyer3, Sudhakar Sivasubramaniam1*
1Department of Biotechnology, Manonmaniam Sundaranar University, Tirunelveli, Tamilnadu- 627012, India
2Centre for Cardiovascular Biology and Disease, Institute for Stem Cell Science and Regenerative Medicine (inStem), Bangalore, India
3Division of Entomology, Centre of Advanced Study in Life Sciences, Manipur University, Imphal-795003, India
4Department of Genetics, School of Biological Sciences, Madurai Kamaraj University, Madurai, Tamilnadu- 625021, India
5Department of Biotechnology, Kalasalingam Academy of Research and Education, Krishnankoil, Tamilnadu-626126, India
6Centre for Information Technology & Engineering, Manonmaniam Sundaranar University, Tirunelveli, Tamilnadu- 627012, India
7Department of Statistics, Manipur University, Imphal – 795 003, India
*Corresponding Author: Sudhakar Sivasubramaniam
Department of Biotechnology, Manonmaniam Sundaranar University, Tirunelveli, Tamilnadu- 627012, India
Tel: 9940998936
Fax: 04634-283270
E-mail: sudhakar@msuniv.ac.in
Received: July 19, 2021 Accepted: August 24, 2021 Published: August 31, 2021
Citation: Paul S, Balakrishnan S, Arumugaperumal A, Thangapandi EJJSB, Devi HS, et al. (2021) Genome sequencing, Analysis and Characterization of Baculovirus Infecting the Caterpillar, Spilosoma obliqua Walker (Arctiidae) (Insecta: Lepidoptera) from India. J Virol Antivir Res 10:5.
Abstract
The Bihar hairy caterpillar, Spilosoma obliqua is an economically important polyphagous insect and to reduce its pest density, baculoviruses are considered as the ideal eco-friendly pathogens under the pest management program. A nucleopolyhedrovirus infecting Spilosoma obliqua (SpobNPV) has been found to be a promising pathogen in the present study. Our report describes the pathogenicity, structural details and genome sequence characterization of SpobNPV-Manipur isolate. The pathogenicity of the virus was studied in terms of median lethal concentration (LC50) and survival time (ST50) and the LC50 of SpobNPV on third instar larvae was 2.7 x 105 POBs/ml and the median survival time (ST50) was 144 hours. The occlusion bodies (OBs) of the virus were purified and the viral genome was sequenced, annotated and compared with other baculoviruses. The sequenced genome of SpobNPV-Manipur isolate was 136,306 bp in length with GC content of 44.9% and it comprises a total of 144 ORFs. The gene content analysis suggested the presence of 13 SpobNPV genes associated with replication, 12 genes associated with transcription and 31 structure related genes. The pathogenicity, structural information and genome resources of SpobNPV-Manipur isolate virus can be utilized further to understand its molecular and genetic mechanisms and improve its efficacy in pest management through recombinant DNA technology.
Keywords: Pest-management; Spilosoma obliqua; Baculovirus; Pathogenicity; Genome sequence
Introduction
The Bihar Hairy Caterpillar (BHC), Spilosoma obliqua Walker (Lepidoptera: Arctiidae) is an important polyphagous pest infesting grams, oilseeds, Jute and pulse crops [1-3]. It has been estimated that S. obliqua could reduce the yield to 77% on soybeans [4]. It is a multivoltine species, emerges in large numbers during March after a long resting phase in winter and maintains appreciable density during June–August that coincides with the availability of their summer hosts [5]. In order to combat the pest problem in general, the application of viral pesticide has been found to be one of the effective and eco-friendly methods [3,6,7]. Under natural conditions, S. obliqua was found infected with nucleopolyhedrovirus (NPV) at the experimental site within the premises of Manipur University [1]. The NPV infecting S. obliqua was first reported from Kerala, in India [8,9]. The bioassay, EM studies of polyhedra and genome analysis of Spilosoma obliqua nucleopolyhedrovirus (SpobNPV) (isolates from Delhi, Kerala and Nagpur-IIPR) were attempted by several researchers [10- 14]. Recently, the draft genome of the SpobNPV virus has been sequenced and deposited in NCBI (KY550224) by Akram et al., (Unpublished work), but the in-depth analysis viz., annotation and characterization of the genome of the Manipur isolate were not studied by them. The viral genome features of SpobNPV-Manipur isolate (Manipur University Campus 24.7475 ºN–93.9370ºE) has not been explored so far. Therefore, the present paper focuses on details pertaining to LC50, ST50, EM studies of polyhedral occlusion bodies (POBs), genome sequencing, annotation and characterization of SpobNPV-Manipur isolate. Further, details pertaining to the above new isolate would enable us to formulate suitable eco-friendly viral pesticides to combat S. obliqua which is a serious pest at the foothills of the Eastern Himalaya and adjoining terrains of north-eastern India, especially in certain parts of Indo- Myanmar hotspot region.
Materials and Methods
Insect rearing
The field-collected egg clusters of Bihar hairy caterpillar, S. obliqua were reared in the entomology laboratory at Manipur University by keeping the eggs in Petri-plates (8.5 cm diameter) over a moistened filter paper. Newly hatched larvae were transferred to fresh leaves inside the conventional insect cage (15 x I5 x 15 cm) with the help of a moist camel hairbrush. The first three larval instars were reared in the above-said cage, while the later instars were reared in a big cage (30 x 30 x 30 cm). The identity of the insect was established with the help of experts of the Indian Agricultural Research Institute (IARI), New Delhi. A continuous nucleus culture was maintained individually in the lab by rearing S. obliqua on castor foliage at 28 ± 1.5ºc 65 ± 5% RH and 12: 12 L: D cycle.
Preparation and purification of the virus inoculums
The initial inoculum of SpobNPV was isolated from an infected 4th instar larva of S. obliqua collected from the fencing plant, Ipomoea cornea surrounding the vegetable garden within the University Campus, Manipur (24.7475 ºN – 93.9370ºE). The putrefied larva of S. obliqua infected with NPV was homogenized and the occlusion bodies (OBs) were filtered through a double-layered muslin cloth. The filtrate was spun at 112 (x g) for 5 min to remove the pellet having larval cells, tissues and other debris. The supernatant was then centrifuged at 9503 (x g) for 20 min. The Polyhedral occlusion bodies (POBs) pellets were collected after discarding the supernatant and they were resuspended in an appropriate volume of distilled water and stored at 4°C [15,16]. The POBs were counted using Helber bacteria haemo-cytometer (0.02 mm depth). A stock solution of SpobNPV-Manipur isolate with the strength of 3 x 1010 was prepared for bioassay studies by scaling up first prior to the bioassay. The stock concentration of virus i.e. 3 x 1010 was diluted to 3 x 109, 3 x 108, 3 x 107, 3 x 106, 3 x 105, 3 x 104 and 3 x 103 POBs/ml by serial dilution method.
Median Lethal Concentration
To assess the median lethal concentration - LC50, uninfected normal healthy 3rd instar caterpillars of S. obliqua were taken from the lab-reared nucleus culture and were inoculated individually with different concentrations ranging from 103 to 109 POBs per ml of the polyhedral of SpobNPV by conventional leaf disc contamination method [17]. To inoculate the caterpillars, S. obliqua was reared on the castor leaf. There were seven concentrations (seven treatments); in addition to control and 30 individuals were tested in each treatment for each insect species. In this method, the field-collected fresh young leaf was cut into a circular fashion (4 cm diameter) and each leaf disc was applied with 100 μl of the virus inoculums with the help of a micropipette and spread on the abaxial and adaxial surface of the leaf with a small tuft of 5-6 hairs of the camelin brush (0-number) and on-air drying, the leaf disc was kept individually inside the petri plate. The pre-starved (for 5 hours) 3rd instar larvae collected from the stock culture was then released individually on the leaf disc. On feeding the virus applied leaf disc, the larvae were reared inside the insect cage for each treatment separately on castor foliage. Care was taken to remove the excreta periodically and fresh leaves were provided daily. The larval mortality rate was noted every day and it was subjected to Abbott’s formula [18] to get corrected percent mortality.
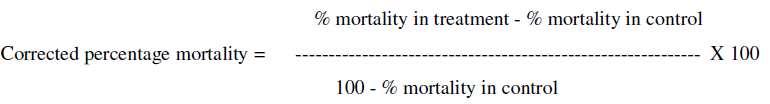
The mortality data thus obtained were subjected to probit analysis [19]. The LC50 and fiducial limits were calculated for each treatment and the regression lines were plotted and slopes of the regression were determined. To calculate the median survival time (ST50), the third instar larvae (n=30 for each species) were inoculated with LC50 dose of POBs obtained in the lethal concentration studies. After inoculation, the caterpillars were reared as stated above and the mortality rate was noted every day. The relation between time factor and larval mortality was processed for Kaplan-Meier Estimate [20] using the software SPSS version 25.
Purification of virus and extraction of viral genomic DNA
The polyhedral inclusion bodies (PIBs) were purified according to the protocol of [21]. The purified PIBs were lysed in 0.1 M Na2CO3 and incubated with proteinase K (microgram/ml) at 50°C for overnight [15,22]. The viral genomic DNA was isolated by using the DNeasy kit (Qiagen, Hilden, Germany) as per the manufacturer’s protocol. The quantitative and qualitative analysis of the extracted DNA was performed by using the NanoDrop2000 (Thermo Scientific, Waltham, USA) and Agarose gel (1%) electrophoresis, respectively.
Genome sequencing, assembly and annotation of the genome contigs
Viral genome sequencing was performed using the Ion Torrent Personal Genome Machine (Life Technologies, Carlsbad, CA). The quality of the Ion Torrent generated raw reads was analysed by using the FastQC quality control tool version 0.11.8 (https://www. bioinformatics.babraham.ac.uk/projects/fastqc/) and CLC genomics workbench version 12.0 [23,24]. The adapter sequences and lowquality reads were filtered by using the trim settings as trim using quality score limit: 0.05; trim ambiguous nucleotides: maximal 2 nucleotides allowed. The filtered reads of the virus were assembled by using the de novo assembly algorithm of CLC genomics workbench version 12.0 with parameters: word size: 20; bubble size: 50 and minimum contig length: 200. The genome coverage statistics for the SpobNPV virus was obtained by mapping the genomic reads to the assembled contigs using the parameters: mismatch cost: 2, insertion cost: 3, deletion cost: 3, length fraction: 0.5 and similarity fraction 0.8. Simultaneously we also opted for reference-based assembly of the SpobNPV (Spilosoma obliqua NPV) genome by using the previously reported Spilosoma obliqua nucleopolyhedrovirus (SpobNPV) isolate IIPR genome as a reference in the CLC genomics workbench. The mapped reads obtained from the reference-based assembly were used to detect all the SNPs, InDels and other structural variations between the two genomes by using the Basic Variant Detection tool and the InDels and Structural Variants tool of CLC genomics workbench. The ORFs were predicted by using the Glimmer (Gene Locator and Interpolated Markov ModelER) prokaryotic gene prediction software integrated within the OmicsBox version 1.1. The ATG initiated ORFs of 50 codons or larger with minimal degree of overlap (<25 codons or <75 nt) were taken into consideration [25]. The Glimmer predicted SpobNPV ORFs were identified from the BLAST search against the NCBI virus database using the BLASTx algorithm [26] with E-value threshold 1E-05. The graphical circular maps of the SpobNPV genome describing the sequence feature, GC contents and annotated gene details were created by using Glimmer predicted GFF (general feature format) annotation files in Geneious Prime sequence analysis software version 2019.1 [27]. The circular genome comparison between our Spilosoma obliqua NPV (SpobNPV) and the reported Spilosoma obliqua nucleopolyhedrovirus isolate IIPR genomes were performed by using the BLAST Ring Image Generator (BRIG) tool version 0.95 [28] with the parameters like alignment algorithm: BLASTn; upper identity threshold: 70%; lower identity threshold: 50% and ring size: 30. The early and late promoter motifs in the SpobNPV virus were detected as described previously [29] by using the neural network promoter prediction tool. The early promoter motifs were defined as TATA and CAGT sequences and the late promoter motif was considered as the TAAG sequence [30,31]. The gene parity plot was carried out by using SpobNPV-Manipur isolate ORFs number as the X-axis and other baculoviruses ORFs as the Y-axis [32]. The scatter plot was drawn by using the Graph Pad Prism software, version 8.2.1.
Phylogenetic analysis with other baculoviruses
The nucleotide sequences of 38 core genes from SpobNPVManipur isolate and other 79 baculoviruses were obtained for phylogenetic analysis [33]. The sequences were concatenated using the Geneious Prime sequence analysis software version 2019.1. The concatenated nucleotide sequences were aligned by using the ClustalW multiple sequence alignment tool with the parameters: gap opening penalty: 10; gap extension penalty: 0.2; protein weight matrix: BLOSUM and gap separation distance: 4 [34]. The phylogenetic tree was constructed through the maximum likelihood method with 100 bootstrap replicates and Kimura’s two-parameter (K2P) nucleotide substitution model using the MEGA 7 software [35,36].
Multiple genome alignment and phylogenomic analysis of SPobNPV v genome
The assembled sub-genomic contigs of SpobNPV was concatenated using the Geneious Prime sequence analysis software version 2019.1. The concatenated genome sequence of SpobNPV virus was aligned to the genomic dataset of its neighboring alphabaculovirus species: Antheraea pernyi nucleopolyhedrovirus (NC_008035), Choristoneura fumiferana multiple nucleopolyhedrovirus (NC_004778), Choristoneura murinana nucleopolyhedrovirus (NC_023177), Choristoneura occidentalis nucleopolyhedrovirus (NC_021925), Choristoneura rosaceana nucleopolyhedrovirus (NC_021924), Hyphantria cunea nucleopolyhedrovirus (NC_007767), Orgyia pseudotsugata multiple nucleopolyhedrovirus (OpMNPV), Philosamia cynthia ricini nucleopolyhedrovirus (JX404026) and Spilosoma obliqua nucleopolyhedrovirus isolate IIPR (SpobNPV IIPR) (KY550224) (Akram et al., 2018 (Unpublished work)) by using the Mauve multiple genome alignment tool (http://darlinglab. org/mauve/mauve.html) [37]. The Average Nucleotide Identity (ANI) score of the SpobNPV-Manipur isolates to its closely related baculovirus genomes were determined by using the Orthologous Average Nucleotide Identity Tool (OAT) (https://www.ezbiocloud. net/tools/orthoani) [38]. The phylogenomic analysis of the NPV virus genomes was carried out by using the REALPHY phylogeny builder server 1.2 [39] and the phylogenomic tree was reconstructed through MEGA 7 software.
Result and Discussion
Pathogenicity of the viral isolates and the structure of PIBs
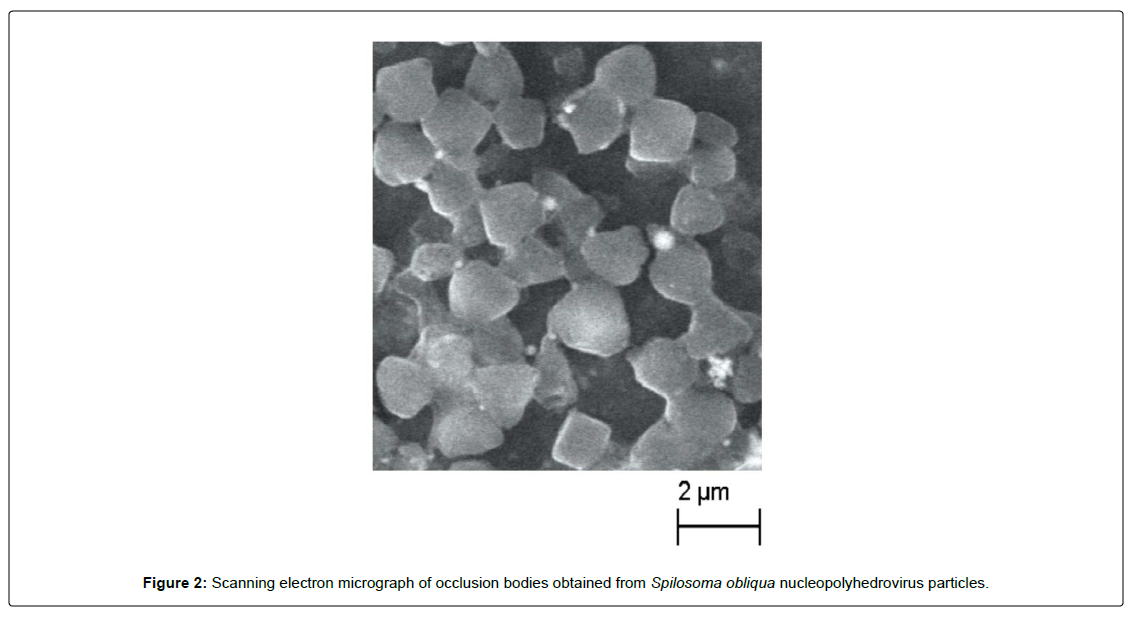
The caterpillars of S. obliqua have five larval instars and the total larval duration ranged from 28 to 32 days when reared at different temperatures and host plants [1]. They exhibit voracious feeding habits especially during the 5th instar (Figure 1A). Therefore, considering their feeding propensity, the suppression of the caterpillar population becomes essential during their early stages. Keeping this view in mind, lab-bioassays of SpobNPV was carried out against 3rd instar larvae of S. obliqua intending to find out the median lethal concentration (LC50) of the pathogen concerned. The result of the experiment revealed that the LC50 of SpobNPV on 3rd instar larvae was 2.7 x 105 POBs/ml (Table 1) and the median survival time (ST50) was 144 hours respectively (Table 2). Earlier studies pertaining to bioassay of SpobNPV revealed that the LC50 value for the 4th instar larva was 5 x 105 POBs/ml [13] and 4.37 x 103 POBs/ml for the 3rd instar larva when inoculated with SpobNPV isolate from Kerala [14]. The Delhi isolate of SpobNPV showed the LC50 value of 4.9 x 102; 2.5 x 104 and 3.16 x 105 POBs/ml respectively for the 2nd, 3rd and 4th instar larvae of S. obliqua [10]. While making a comparative analysis on the LC50 value of three different isolates of SpobNPV from three different geographical zones such as Kerala (Southern India), Delhi (Northern India) and Manipur (North-Eastern India), the study reflected that the LC50 was respectively 4.37 x 103, 3.16 x 105 and 2.7 x 105 POBs/ ml [10,14]. The median survival time (ST50) was 181 and 167 h respectively, at a dose of 1 x 106 and 1x 108 POBs/ml of SpobNPV. Based on the LC50, it is evident that the isolate from Kerala appeared to be more virulent than that of Manipur isolate of the present study. The infected larvae exhibited typical symptoms such as restless movement, shiny and lose cuticle, oozing out hemolymph from the oral end (Figure 1B). Their food intake reduced drastically from 96 h of p.i. However, a few larvae showed cannibalistic behaviour too. At the advanced stage of post-infection (p. i), they tend to move towards the apical portion of the twig invariably and hang upside down with their caudal legs (Figure 1B). Each larva can be said to be a bioreactor by virtue of producing as much as 1012 POBs when inoculated with 104 POBs. The occlusion bodies of SpobNPV (Manipur isolate) were purified and the morphology was studied by scanning electron microscope (Figure 2). The average size of the occlusion bodies of SpobNPV was 2.351 ± 0.857 μm. The occlusion bodies were found to be tetrahedral shape similar to the POBs of Spilarctia obliqua NPV reported in the Kumar et al., [14].
| Insect Virus (Pathogen) | Larval host (third instar) | Regression Equation | LC50 (POBs/ml) | Fiducial limit@ (upper & lower) |
|---|---|---|---|---|
| X2 | ||||
| SpobNPV | Spilosoma obliqua Walker (Arctiidae) (n = 210) |
Y = 0.41x + 2.36 | 2.7 x 105 | 1.4 x 106 1.6 x 105 |
| 14.95 ** | ||||
| **Heterogenous; $(Probit Analysis – Finney, 1971); @ Fiducial limit at 95% confidence level. N–represents number of larvae tested against Spilosoma obliqua insect virus. All the larvae were reared at 28 ± 1.5º; 65± 5% RH & 12:12 L: D |
||||
Table 1: Median Lethal Concentration$ (LC50) of Spilosoma obliqua insect virus against its lepidopteran host.
| Insect Virus (Pathogen) | Larval host (third instar) | ST50 (hours) | Standard Error | Fiducial limit@ (upper & lower) |
|---|---|---|---|---|
| SpobNPV | Spilosoma obliqua (Arctiidae) | 144 | 18.7 | 180.7 – 107.3 |
| Overall comparison based on Kaplan-Meier Estimate: Log Rank (Mantel-Cox) X2 = 6.614, df = 1, P – value = 0.010. (Upper and lower limits @ 95% confidence level) ST50 value was arrived at by inoculating the caterpillar (n=50) with the LC50 concentration of viral pathogen mentioned in Table-1 for the concerned host larvae. | ||||
Table 2: Median Survival Time (ST50) of Spilosoma obliqua insect virus against its lepidopteran host.
The striking feature of S. obliqua is that they occur in a cluster under the field condition at least up to 3rd instar stage and hence, they occur in a localized group during the early period of infestation. Therefore, giving a drenching spray of viral pesticide during the early stage of the caterpillar not only make them susceptible but prevents further dissemination of the caterpillars from infested plant to a healthy plant. Field evaluation studies have unambiguously revealed that spraying with 250 larval extracts of virus-infected larvae (≈5 x 1012 POBs/ml) per h could reduce infestation by the S. obliqua substantially [40].
Genome sequencing, quality control and assembly
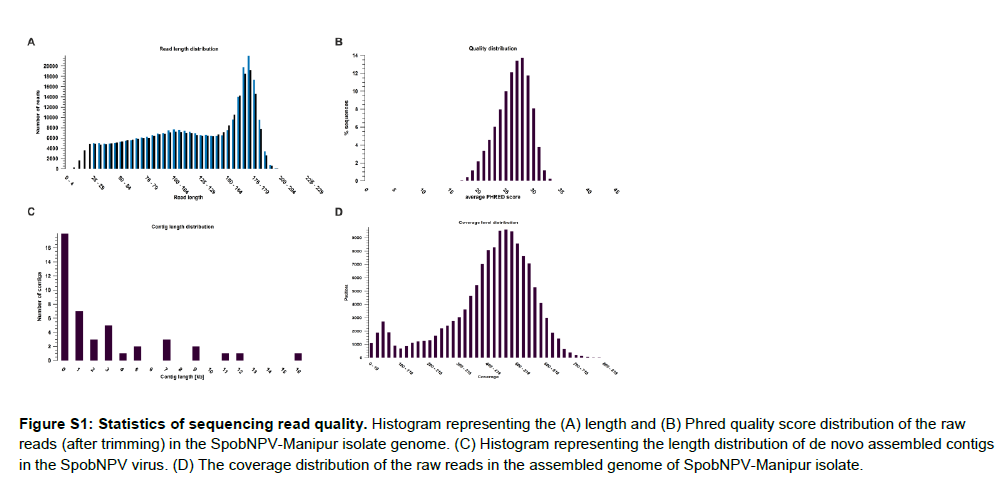
The whole-genome sequencing of the SpobNPV virus using Ion Torrent personal genome machine (Life Technologies, Carlsbad, CA) generated a total of 535,029 reads with an average length of 114.8 bp. The FastQC (version.0.11.5) quality assessment software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [23] and CLC genomics workbench version 12.0 [24] were used to analyze the read quality and trim the ambiguous, low quality reads from the genome dataset. A total of 148,616 reads were trimmed based on their quality and the average length of the final filtered reads was obtained as 112 bp (Figure S1A and Table 3). The Phred quality distribution of the cleaned reads suggested that 98.16% of SpobNPV reads had average quality PHRED-score of 20 and above (Figure S1B). The de novo assembly of the trimmed filtered reads using the CLC genomics workbench version 12.0 generated a total of 41 contings with an average length of 3,266 bp and total genome size of 136.3 kb. The summary statistics of the CLC assembly denoted that the N75 value, N50 value and GC% for the assembled genome were 4,684 bp, 7,508 bp and 44.9% respectively (Table 3). The genome sequence reads were deposited to the NCBI sequence read archive (SRA) with the Accession: SRX6949976 the BioProject ID- PRJNA560447. Among the 41 genomic contigs of SpobNPV, 22 contigs had sequence length larger than 1,000 bp and the largest contig length was 16,279 bp (Figure S1C).
| Summary | SpobNPV |
|---|---|
| No. of Raw Reads | 535,029 |
| Average length of Raw Reads (bp) | 114.8 |
| No. of Reads trimmed | 148,616 |
| Average length of Clean Reads (bp) | 112 |
| Total No. of Contigs | 41 |
| Max Contig length (bp) | 16,279 |
| Min Contig length (bp) | 165 |
| Mean Contig length (bp) | 3,266 |
| N75 length (bp) | 4,684 |
| N50 length (bp) | 7,508 |
| GC% | 44.9 |
| Size of genome (bp) | 136,306 |
Table 3: Summary statistics of SpobNPV genome assembly.
The de novo assembled genome size and GC content of the SpobNPV-Manipur isolate was found to be reasonably higher compared to the reported group I alphabaculoviruses like Bombyx mandarina nucleopolyhedrovirus (BomaNPV) [41], Bombyx mori nucleopolyhedrovirus (BmNPV) [42] etc. The genome coverage summary statistics denoted that a total of 508,000 SpobNPV reads were mapped to their assembled genome with an average coverage of 424.65 (Table S1). Besides, the coverage level distribution represented that 99.7% of the SpobNPV genome had coverage between 1 and 1,104 (Figure S1D). In Parallel, the reference-based assembly using Spilosoma obliqua nucleopolyhedrovirus isolate IIPR genome (KY550224) as reference generated 136.1 kb genome for our reported SpobNPV virus with the GC content of 45.4% and average coverage of 219.25 (File S1). The mapped reads obtained from the referencebased assembly demonstrated a total of 465 variants between the genomes of our SpobNPV-Manipur isolate and reported Spilosoma obliqua nucleopolyhedrovirus (SpobNPV) isolate IIPR. Among these 465 variants, 261 were SNVs (single nucleotide variants), 14 were MNVs (multi nucleotide variants) and 190 were InDels and structural variants. Of the 190 InDels and structural variants, a total of 28 deletions, 114 insertions, 9 inversions, 20 replacements and 19 other structural variants were observed between the two genomes (Table S2). The list of all the SNVs and MNVs was documented in Table S3 [43].
Annotation and characterization of SpobNPV genome
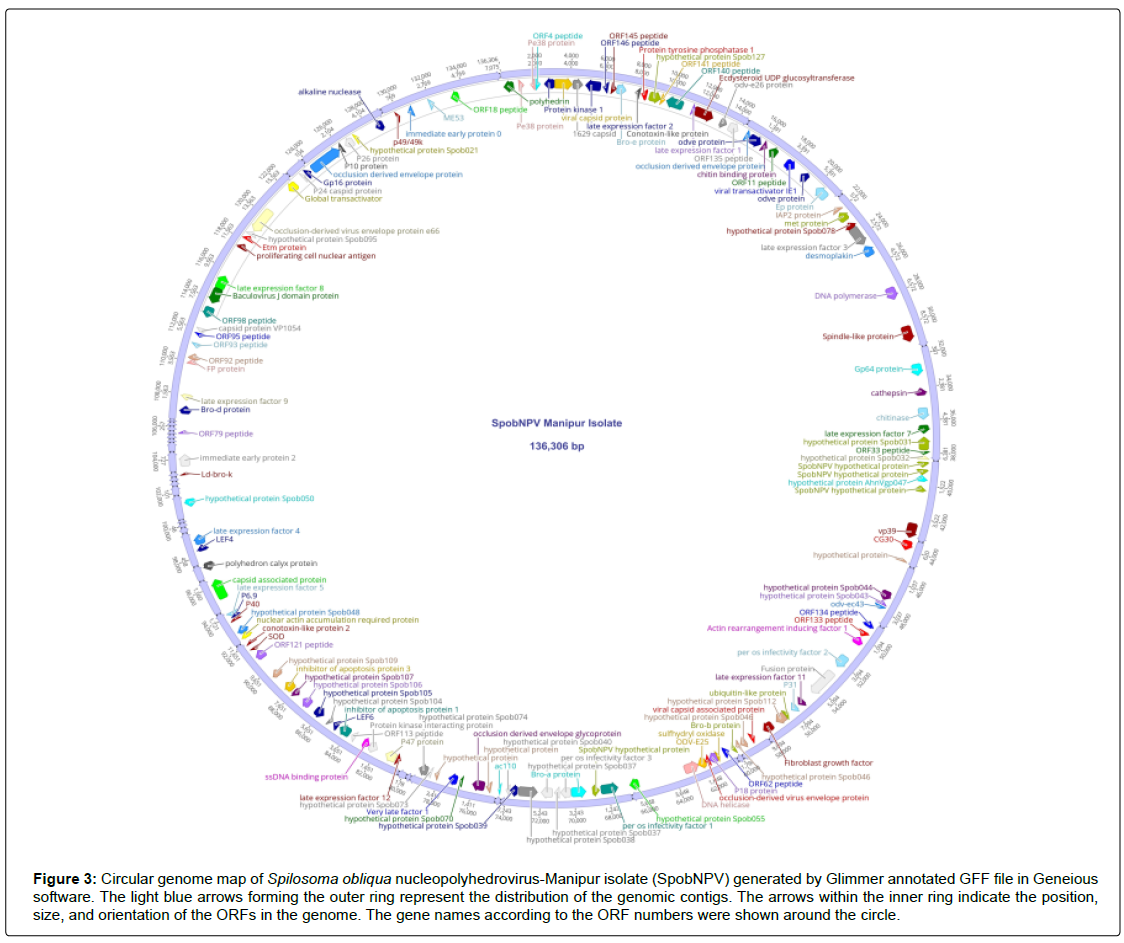
The genomic contigs of the SpobNPV virus were annotated by using the Glimmer (Gene Locator and Interpolated Markov ModelER) prokaryotic gene finding tool within OmicsBox version 1.1 (https://www.biobam.com/omicsbox/). A total of 144 ORFs code for proteins >50 amino acids were predicted from Glimmer annotation. The polyhedrin gene with reverse orientations was considered as ORF 1 according to the convention [44]. The successive nucleotides were numbered according to the orientation of the polyhedrin gene. A total of 88 (61.1%) ORFs were present in the forward orientation and 56 (38.8%) SpobNPV ORFs were present in the reverse orientation in the genome map (Figure 3). The distribution of orientations for the SpobNPV ORFs was uneven like the reported gammabaculovirus Neodiprion sertifer nucleopolyhedrovirus (NeseNPV) [30]. The SpobNPV genome sequences were scanned for promoter motifs within 300 bp upstream of the start codon for each ORFs through neural network promoter prediction [45]. The genome-wide promoter scanning detected that 59 SpobNPV ORFs have baculovirus early promoter motifs (TATA box and CAGT motif sequence), 13 ORFs have late promoter motifs (TAAG), 39 ORFs contain both the motifs and 33 ORFs lack any consensus promoter sequences in their upstream region.
Figure 3: Circular genome map of Spilosoma obliqua nucleopolyhedrovirus-Manipur isolate (SpobNPV) generated by Glimmer annotated GFF file in Geneious software. The light blue arrows forming the outer ring represent the distribution of the genomic contigs. The arrows within the inner ring indicate the position, size, and orientation of the ORFs in the genome. The gene names according to the ORF numbers were shown around the circle.
The Glimmer predicted ORFs were subjected to BLAST annotation against NCBI baculovirus database using the OmicsBox version 1.1 and classified according to their homologous sequences they aligned with the highest bit score. The detailed list of the functionally annotated genes for the SpobNPV genome dataset was documented in Table 4. Among the 144 SpobNPV ORFs predicted from Glimmer annotation, 140 ORFs were homologous to at least one other baculovirus. Besides, the Glimmer annotation predicted 38 baculovirus core genes and 21 lepidopteran baculovirus conserved genes in the SpobNPV genome (Table 5). Notably, the core gene p6.9 was absent in the genome of Spilosoma obliqua nucleopolyhedrovirus isolate IIPR (SpobNPV IIPR) (NCBI GenBank accession: KY550224). Our study provides the first report regarding the presence of this core gene in the genome dataset of the SpobNPV virus.
| Name | ORF number | ORF starts | ORF ends | Strand | Promoter | Length (aa) | ORF position | Amino acid identity (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SpobNPV IIPR | HycuNPV | PhcyNPV | CfMNPV | SpobNPV IIPR | HycuNPV | PhcyNPV | CfMNPV | |||||||
| polyhedrin | 1 | 66 | 638 | - | 190 | 1 | 1 | 1 | 1 | 100% | 100% | 98% | 98.33% | |
| Pe38 protein | 2 | 953 | 1264 | + | E,L | 103 | 5 | 5 | 5 | 144 | 100% | 100% | 76% | 68.42% |
| Pe38 protein | 3 | 1623 | 1898 | - | E | 91 | 5 | 5 | 5 | 144 | 100% | 100% | 80% | 68.42% |
| ORF4 peptide | 4 | 1973 | 2218 | + | L | 81 | 4 | 100% | ||||||
| Protein kinase 1 | 5 | 2450 | 3031 | - | E | 193 | 3 | 3 | 3 | 145 | 93% | 93.78% | 83% | 87.56% |
| viral capsid protein | 6 | 3033 | 4109 | + | E | 358 | 2 | 2 | 2 | 146 | 90% | 88.68% | 62% | 72.32% |
| 1629capsid | 7 | 4135 | 4692 | + | 185 | 2 | 2 | 2 | 146 | 100% | 100% | 65% | 75% | |
| late expression factor 2 | 8 | 4869 | 5768 | - | E | 299 | 132 | 147 | 137 | 3 | 99% | 99.35% | 79% | 81.82% |
| ORF146 peptide | 9 | 5925 | 6191 | - | 88 | 131 | 146 | 4 | 100% | 100% | 72.22% | |||
| ORF145 peptide | 10 | 6427 | 6696 | + | E | 89 | 130 | 145 | 136 | 5 | 100% | 100% | 85% | 82.02% |
| Bro-e protein | 11 | 6750 | 7259 | + | 169 | 129 | 144 | 98% | 98.22% | |||||
| Conotoxin-like protein | 12 | 7837 | 8100 | - | L | 53 | 143 | 133 | 131 | 100% | 96% | 88.68% | ||
| Protein tyrosine phosphatase 1 | 13 | 8188 | 8526 | - | 112 | 128 | 142 | 132 | 9 | 100% | 100% | 83% | 86.92% | |
| hypothetical protein Spob127 | 14 | 8709 | 9389 | + | E | 226 | 127 | 141 | 131 | 10 | 100% | 100% | 70% | 72.43% |
| ORF141 peptide | 15 | 9415 | 9612 | + | E | 65 | 127 | 141 | 131 | 10 | 92% | 92.42% | 63% | 64.71% |
| ORF140 peptide | 16 | 9713 | 10714 | - | E | 333 | 126 | 140 | 129 | 12 | 98% | 99.7% | 84% | 84.19% |
| late expression factor 1 | 17 | 11348 | 11500 | + | 50 | 125 | 139 | 127 | 13 | 100% | 100% | 100% | 86% | |
| Ecdysteroid UDP glucosyltransferase | 18 | 11488 | 12654 | + | 388 | 124 | 138 | 126 | 14 | 99% | 99.71% | 89% | 81.02% | |
| odv-e26 protein | 19 | 13153 | 13548 | + | E,L | 131 | 123 | 137 | 125 | 15 | 96% | 100% | 63% | 75.59% |
| ORF135 peptide | 20 | 13711 | 14340 | + | E,L | 209 | 122 | 135 | 124 | 16 | 100% | 100% | 80% | 83.65% |
| occlusion derived envelope protein | 21 | 14854 | 15126 | + | E,L | 90 | 13 | 14 | 14 | 136 | 100% | 100% | 84% | 86.67% |
| odve protein | 22 | 15150 | 15911 | + | E,L | 253 | 12 | 13 | 13 | 137 | 100% | 100% | 92% | 94.67% |
| chitin binding protein | 23 | 16053 | 16340 | + | E,L | 95 | 11 | 12 | 12 | 138 | 100% | 100% | 88% | 93.68% |
| ORF11 peptide | 24 | 16490 | 16996 | - | E | 168 | 10 | 11 | 11 | 139 | 97% | 97.37% | 79% | 86.18% |
| viral transactivator IE1 | 25 | 17763 | 18341 | + | E,L | 192 | 9 | 10 | 10 | 140 | 100% | 100% | 78% | 93.29% |
| odve protein | 26 | 18884 | 19369 | - | E,L | 161 | 8 | 9 | 9 | 141 | 99% | 99.38% | 87% | 96.15% |
| Ep protein | 27 | 20148 | 20915 | - | E | 255 | 6 | 8 | 100% | 94.12% | ||||
| IAP2 protein | 28 | 21741 | 22040 | - | E | 99 | 76 | 80 | 77 | 66 | 100% | 100% | 74% | 91.01% |
| met protein | 29 | 22178 | 22753 | - | 191 | 77 | 81 | 78 | 65 | 100% | 98.43% | 84% | 86.39% | |
| hypothetical protein Spob078 | 30 | 22947 | 23354 | - | E | 135 | 78 | 82 | 79 | 64 | 100% | 97.67% | 91% | 92.31% |
| late expression factor 3 | 31 | 23356 | 24570 | + | E,L | 404 | 79 | 83 | 80 | 63 | 99% | 99.19% | 75% | 86.83% |
| desmoplakin protein | 32 | 24697 | 25338 | - | E | 213 | 80 | 84 | 62 | 100% | 96.68% | 64.71% | ||
| DNA polymerase | 33 | 27608 | 28354 | + | 248 | 81 | 85 | 82 | 61 | 100% | 100% | 88% | 90.61% | |
| Spindle-like protein | 34 | 30161 | 31120 | + | E,L | 319 | 82 | 86 | 83 | 60 | 99% | 99.69% | 79% | 75.14% |
| Gp64 protein | 35 | 32552 | 33268 | + | 238 | 27 | 28 | 30 | 119 | 100% | 98.72% | 94% | 96.6% | |
| cathepsin | 36 | 34039 | 34476 | - | E | 145 | 28 | 29 | 118 | 98% | 97.73% | 88.89% | ||
| chitinase | 37 | 35256 | 35948 | + | E | 230 | 29 | 30 | 117 | 93% | 93.01% | 91.98% | ||
| late expression factor 7 | 38 | 36311 | 36820 | + | E,L | 169 | 30 | 31 | 31 | 115 | 90% | 90.85% | 51% | 72.11% |
| hypothetical protein Spob031 | 39 | 37035 | 37829 | - | E | 264 | 31 | 32 | 33 | 113 | 99% | 98.77% | 77% | 81.25% |
| ORF33 peptide | 40 | 37912 | 38115 | + | E | 67 | 33 | 34 | 112 | 98.51% | 60% | 76% | ||
| hypothetical protein Spob032 | 41 | 38154 | 38399 | - | E | 81 | 32 | 34 | 35 | 111 | 100% | 97.53% | 79% | 80.25% |
| SpobNPV hypothetical protein | 42 | 38556 | 38867 | + | E,L | 103 | ||||||||
| SpobNPV hypothetical protein | 43 | 39013 | 39351 | + | E | 112 | ||||||||
| hypothetical protein AhnVgp047 | 44 | 39369 | 39743 | - | E | 124 | 91 | 50% | ||||||
| SpobNPV hypothetical protein | 45 | 40038 | 40373 | - | E | 111 | ||||||||
| vp39 | 46 | 42362 | 43284 | + | E, L | 205 | 61 | 65 | 65 | 81 | 99.66% | 95.96% | 76.41% | 81.67% |
| CG30 | 47 | 43478 | 44038 | + | E | 186 | 62 | 66 | 66 | 80 | 100% | 100% | 69% | 84.32% |
| hypothetical protein | 48 | 44586 | 44747 | - | E | 53 | 67 | 87% | ||||||
| hypothetical protein Spob044 | 49 | 46787 | 47359 | + | L | 190 | 44 | 48 | 46 | 98 | 100% | 93.16% | 75% | 76.32% |
| hypothetical protein Spob043 | 50 | 47443 | 47664 | - | 73 | 43 | 47 | 45 | 99 | 98% | 97.01% | 90% | 88.06% | |
| odv-ec43 | 51 | 47764 | 47964 | + | E | 66 | 42 | 46 | 44 | 100 | 98.75% | 98.75% | 78.75% | 82.50% |
| ORF134 peptide | 52 | 48909 | 49256 | - | 115 | 121 | 134 | 123 | 17 | 100% | 100% | 75% | 81.82% | |
| ORF133 peptide | 53 | 49611 | 49925 | + | L | 104 | 120 | 133 | 122 | 18 | 100% | 100% | 78% | 84.62% |
| Actin rearrangement inducing factor 1 | 54 | 50042 | 50518 | - | E | 158 | 119 | 132 | 19 | 100% | 100% | 57.25% | ||
| per os infectivity factor 2 | 55 | 51491 | 52288 | + | E | 265 | 118 | 131 | 120 | 20 | 98% | 98.44% | 95% | 95.18% |
| Fusion protein | 56 | 52742 | 54457 | + | 571 | 117 | 130 | 119 | 21 | 100% | 100% | 70% | 75.17% | |
| late expression factor 11 | 57 | 55170 | 55511 | + | E | 113 | 115 | 128 | 23 | 100% | 100% | 87.5% | ||
| P31 | 58 | 55716 | 56057 | + | E | 113 | 114 | 127 | 117 | 24 | 100% | 100% | 82% | 84% |
| ubiquitin-like protein | 59 | 56309 | 56548 | - | E | 79 | 113 | 126 | 116 | 25 | 100% | 100% | 96% | 96.1% |
| hypothetical protein Spob112 | 60 | 56565 | 57194 | + | E | 209 | 112 | 125 | 115 | 26 | 100% | 99.52% | 80% | 85.65% |
| Fibroblast growth factor | 61 | 57527 | 58114 | + | E | 195 | 111 | 124 | 114 | 27 | 100% | 100% | 73% | 75.86% |
| viral capsid associated protein | 62 | 58763 | 59029 | - | 88 | 45 | 49 | 97 | 100% | 100% | 63.44% | |||
| hypothetical protein Spob046 | 63 | 59378 | 59737 | + | E | 119 | 46 | 50 | 50 | 96 | 85% | 84.13% | 83% | 81.51% |
| hypothetical protein Spob046 | 64 | 59800 | 60009 | + | E | 69 | 46 | 50 | 50 | 96 | 91% | 91.53% | 81% | 81.36% |
| Bro-b protein | 65 | 60111 | 60350 | + | 79 | 51 | 56 | 96% | 95.08% | |||||
| ORF62 peptide | 66 | 60518 | 60820 | - | E | 100 | 58 | 62 | 62 | 84 | 97% | 97.89% | 65% | 71.58% |
| sulfhydryl oxidase | 67 | 61109 | 61303 | - | E,L | 64 | 58 | 62 | 62 | 84 | 100% | 100% | 96% | 98.28% |
| P18 protein | 68 | 61302 | 61781 | + | E,L | 159 | 57 | 61 | 61 | 85 | 100% | 100% | 94% | 94.97% |
| occlusion-derived virus envelope protein | 69 | 61786 | 62049 | + | E,L | 87 | 56 | 60 | 60 | 86 | 98% | 98.77% | 91% | 92.59% |
| ODV-E25 | 70 | 62034 | 62525 | + | E | 163 | 56 | 60 | 60 | 86 | 99% | 100% | 94% | 91.3% |
| DNA helicase | 71 | 62515 | 63375 | - | L | 286 | 54 | 59 | 59 | 87 | 100% | 100% | 95% | 93.73% |
| hypothetical protein Spob055 | 72 | 66200 | 66709 | + | E,L | 169 | 55 | 58 | 58 | 88 | 100% | 99.41% | 94% | 91.67% |
| per os infectivity factor 1 | 73 | 67436 | 68425 | - | E,L | 329 | 33 | 35 | 36 | 110 | 99% | 95.64% | 93% | 94.38% |
| SpobNPV hypothetical protein | 74 | 68536 | 68940 | - | 134 | |||||||||
| Bro-a protein | 75 | 69322 | 70170 | - | E | 282 | 35 | 38 | 38 | 108 | 100% | 92.9% | 82% | 88.17% |
| per os infectivity factor 3 | 76 | 70195 | 70812 | + | E | 205 | 36 | 39 | 39 | 106 | 100% | 94.15% | 83% | 84.24% |
| hypothetical protein Spob037 | 77 | 70889 | 71167 | + | E,L | 92 | 37 | 40 | 40 | 105 | 100% | 95.4% | 70% | 76.74% |
| hypothetical protein Spob037 | 78 | 71287 | 71970 | + | E | 227 | 37 | 40 | 40 | 105 | 98% | 96.89% | 74% | 81.5% |
| hypothetical protein Spob038 | 79 | 72177 | 73265 | - | E | 362 | 38 | 42 | 82 | 104 | 99% | 97.95% | 76% | 86.03% |
| hypothetical protein Spob039 | 80 | 73341 | 73781 | + | E | 146 | 39 | 43 | 41 | 103 | 100% | 97.26% | 75% | 68.52% |
| hypothetical protein Spob040 | 81 | 73821 | 74024 | + | E | 67 | 40 | 44 | 102 | 98% | 94.03% | 82.81% | ||
| ac110 | 82 | 74070 | 74236 | + | E | 55 | 41 | 45 | 101 | 71.43% | 71.43% | 69.64% | ||
| hypothetical protein | 83 | 74914 | 75195 | + | 93 | 67 | 70 | 69 | 76 | 82% | 82.22% | 74% | 92.54% | |
| occlusion derived envelope glycoprotein | 84 | 75314 | 76021 | + | E,L | 235 | 68 | 71 | 70 | 75 | 100% | 100% | 93% | 97.81% |
| hypothetical protein Spob070 | 85 | 76626 | 76805 | + | E,L | 59 | 70 | 73 | 72 | 73 | 90% | 90% | 100% | 94.44% |
| Very late factor 1 | 86 | 76950 | 77459 | + | L | 169 | 71 | 74 | 73 | 72 | 100% | 100% | 91% | 96.3% |
| hypothetical protein | 87 | 78104 | 78301 | + | L | 65 | 72 | 75 | 74 | 71 | 79% | 79.69% | 76% | 76.56% |
| hypothetical protein Spob073 | 88 | 78517 | 78669 | + | L | 50 | 73 | 76 | 75 | 70 | 100% | 100% | 78% | 86% |
| hypothetical protein Spob074 | 89 | 78767 | 79288 | + | E,L | 173 | 74 | 77 | 75 | 69 | 100% | 98.84% | 75% | 79.77% |
| late expression factor 12 | 90 | 80488 | 80733 | - | 81 | 98 | 109 | 100 | 41 | 96% | 96.97% | 73% | 72.37% | |
| P47 protein | 91 | 80788 | 81432 | + | 214 | 99 | 110 | 101 | 40 | 100% | 99.52% | 88% | 86.67% | |
| Protein kinase interacting protein | 92 | 82021 | 82521 | - | L | 166 | 100 | 111 | 102 | 39 | 100% | 100% | 74% | 84.34% |
| ssDNA binding protein | 93 | 82536 | 82928 | - | E | 130 | 101 | 112 | 103 | 38 | 100% | 100% | 90% | 83.85% |
| ORF113 peptide | 94 | 83515 | 83766 | + | E,L | 83 | 102 | 113 | 104 | 37 | 95% | 95.59% | 80% | 79.41% |
| inhibitor of apoptosis protein 1 | 95 | 83894 | 84571 | + | L | 225 | 103 | 114 | 105 | 36 | 100% | 99.4% | 84% | 93.98% |
| LEF6 | 96 | 84721 | 85032 | + | 103 | 115 | 106 | 35 | 100% | 80% | 53.45% | |||
| hypothetical protein Spob104 | 97 | 85148 | 85354 | - | L | 68 | 104 | 116 | 107 | 34 | 100% | 98.53% | 82% | 80.88% |
| hypothetical protein Spob105 | 98 | 85785 | 86354 | - | E,L | 189 | 105 | 117 | 108 | 33 | 99% | 97.79% | 78% | 88.4% |
| hypothetical protein Spob106 | 99 | 86836 | 87342 | + | E | 168 | 106 | 118 | 109 | 32 | 100% | 97.59% | 70% | 72.89% |
| hypothetical protein Spob107 | 100 | 87712 | 88077 | - | 121 | 107 | 119 | 111 | 31 | 99% | 95.73% | 66% | 73.04% | |
| inhibitor of apoptosis protein 3 | 101 | 88225 | 88770 | + | E,L | 181 | 108 | 120 | 105 | 30 | 100% | 99.45% | 48% | 75.39% |
| hypothetical protein Spob109 | 102 | 89383 | 89814 | + | E,L | 143 | 109 | 121 | 100% | 99.3% | ||||
| ORF121 peptide | 103 | 90796 | 91338 | + | E | 180 | 109 | 121 | 100% | 100% | ||||
| SOD | 104 | 91463 | 91633 | - | E,L | 56 | 110 | 122 | 112 | 28 | 100% | 100% | 94% | 98.15% |
| conotoxin-like protein 2 | 105 | 91958 | 92122 | + | L | 54 | 123 | 113 | 62% | 88% | ||||
| nuclear actin accumulation required protein | 106 | 92366 | 92719 | + | E,L | 117 | 47 | 51 | 51 | 95 | 100% | 96.58% | 89% | 84.62% |
| hypothetical protein Spob048 | 107 | 92728 | 93174 | + | E | 148 | 48 | 52 | 52 | 94 | 98% | 97.97% | 81% | 85.81% |
| P40 | 108 | 93617 | 93799 | + | E | 60 | 48 | 52 | 53 | 94 | 100% | 100% | 81% | 94.64% |
| P6.9 | 109 | 93830 | 94006 | + | E,L | 58 | 53 | 54 | 93 | 100% | 58.3% | 77.6% | ||
| late expression factor 5 | 110 | 94003 | 94330 | - | E,L | 98 | 49 | 54 | 55 | 92 | 100% | 100% | 80% | 93% |
| capsid associated protein | 111 | 95122 | 96363 | + | 413 | 65 | 68 | 68 | 78 | 99% | 99.76% | 84% | 87.32% | |
| polyhedron calyx protein | 112 | 97005 | 97496 | - | E,L | 163 | 24 | 25 | 27 | 124 | 100% | 100% | 92% | 93.25% |
| LEF4 | 113 | 98298 | 98651 | + | E,L | 117 | 60 | 64 | 64 | 82 | 100% | 100% | 77% | 86.92% |
| late expression factor 4 | 114 | 98731 | 99291 | + | E | 186 | 60 | 64 | 64 | 82 | 100% | 98.92% | 85% | 88.17% |
| hypothetical protein Spob050 | 115 | 101060 | 101533 | - | E | 157 | 50 | 55 | 56 | 91 | 97% | 97.99% | 81% | 89.86% |
| Ld-bro-k | 116 | 103022 | 103189 | + | 55 | 53 | 57 | 94% | 94.55% | |||||
| immediate early protein 2 | 117 | 103582 | 104268 | + | 228 | 7 | 6 | 8 | 142 | 99% | 98.68% | 70% | 76.19% | |
| ORF79 peptide | 118 | 105555 | 105731 | + | 58 | 79 | 67 | 100% | 82.76% | |||||
| Bro-d protein | 119 | 106766 | 107254 | + | 162 | 52 | 88 | 57 | 100% | 100% | 74% | |||
| late expression factor 9 | 120 | 107581 | 107943 | - | E,L | 120 | 83 | 89 | 84 | 59 | 100% | 100% | 84% | 96.67% |
| FP protein | 121 | 109843 | 110127 | + | E,L | 94 | 85 | 91 | 86 | 57 | 100% | 100% | 91% | 80.85% |
| ORF92 peptide | 122 | 110124 | 110483 | + | E,L | 119 | 86 | 92 | 87 | 56 | 100% | 100% | 76% | 61.8% |
| ORF93 peptide | 123 | 110859 | 111113 | - | 84 | 87 | 93 | 88 | 55 | 98% | 98.77% | 78% | 86.42% | |
| ORF95 peptide | 124 | 111554 | 111757 | - | E | 67 | 88 | 95 | 89 | 53 | 100% | 100% | 76% | 86.89% |
| capsid protein VP1054 | 125 | 111819 | 112088 | - | 89 | 89 | 96 | 90 | 52 | 100% | 100% | 85% | 88.76% | |
| ORF98 peptide | 126 | 112823 | 113476 | - | 217 | 90 | 98 | 91 | 50 | 98% | 98.53% | 84% | 88.97% | |
| Baculovirus J domain protein | 127 | 113771 | 114712 | - | E | 313 | 91 | 101 | 93 | 49 | 99% | 99.65% | 58% | 70.53% |
| late expression factor 8 | 128 | 114739 | 115575 | + | 278 | 92 | 102 | 94 | 48 | 100% | 100% | 89% | 91.61% | |
| proliferating cell nuclear antigen | 129 | 117504 | 117836 | + | E | 110 | 93 | 103 | 131 | 47 | 98% | 95.38% | 57% | 74.67% |
| Etm protein | 130 | 118141 | 118416 | + | E,L | 91 | 94 | 104 | 96 | 46 | 98% | 93.33% | 57% | 90% |
| hypothetical protein Spob095 | 131 | 118518 | 118763 | + | E | 81 | 95 | 105 | 100% | 96.2% | ||||
| occlusion-derived virus envelope protein e66 | 132 | 118825 | 120381 | - | 518 | 96 | 106 | 97 | 45 | 99% | 94.2% | 88% | 89.15% | |
| Global transactivator | 133 | 122022 | 122585 | - | E | 187 | 97 | 108 | 99 | 42 | 100% | 100% | 76% | 85.37% |
| P24 caspid protein | 134 | 123188 | 123379 | + | E | 63 | 26 | 27 | 29 | 122 | 100% | 96.77% | 75% | 77.42% |
| Gp16 protein | 135 | 123462 | 123776 | + | E,L | 104 | 25 | 26 | 28 | 123 | 94% | 93.18% | 81% | 88.64% |
| occlusion derived envelope protein | 136 | 123934 | 125868 | + | 644 | 18 | 19 | 20 | 130 | 100% | 100% | 92% | 93.64% | |
| P10 protein | 137 | 125865 | 126131 | - | E | 88 | 19 | 20 | 21 | 129 | 100% | 98.86% | 95% | 91.76% |
| P26 protein | 138 | 126345 | 126884 | - | E | 179 | 20 | 21 | 22 | 128 | 99% | 99.4% | 85% | 90.18% |
| hypothetical protein Spob021 | 139 | 127064 | 127273 | + | E | 69 | 21 | 22 | 24 | 127 | 87% | 87.1% | 83% | 70.97% |
| alkaline nuclease | 140 | 128428 | 128904 | - | E | 158 | 22 | 23 | 25 | 126 | 92% | 92.68% | 76% | 76.83% |
| p49/49k | 141 | 129822 | 130025 | + | L | 67 | 14 | 15 | 15 | 135 | 100% | 100% | 85.07% | 88.06% |
| immediate early protein 0 | 142 | 130538 | 130822 | - | 94 | 15 | 16 | 16 | 134 | 100% | 100% | 83% | 87.06% | |
| ME53 | 143 | 131892 | 132182 | + | E,L | 96 | 16 | 17 | 17 | 132 | 97% | 97.14% | 77% | 91.43% |
| ORF18 peptide | 144 | 133207 | 133695 | - | E,L | 162 | 17 | 18 | 19 | 99% | 99.38% | 67% | ||
Table 4: SpobNPV-Manipur isolate genome annotation details.
Gene content of SpobNPV-manipur isolate
The genome annotation of the SpobNPV baculovirus demonstrated the presence of 13 replication related genes, 12 transcription-associated genes, 31 structure related genes, 11 genes essentially required for oral infection and 24 auxiliary genes (Table 5). The six genes reported to be essential for the baculovirus DNA replication namely: immediate early gene-1(ie-1), DNA polymerase (DNA-pol), helicase, late expression factor 1 (lef1), lef2 and lef3 [46,47] were present in the genome dataset of SpobNPV-Manipur isolate. Among the other replication-related genes we monitored the presence of lef-7 and pcna. The eukaryotic pcna gene plays a key role in DNA synthesis, repair and progression of cell cycle [48]. In baculovirus AcMNPV the overexpression of pcna stimulates the replication of the viral genome within the host cell. Besides, it also stimulates the transcription of the late genes and enhances the larval mortality rate [49,50]. The late expression gene lef-7 was reported to involve in the transient DNA replication and DNA damage response mechanisms of baculoviruses [51,52]. The presence of these genes indicates their role in regulating the DNA synthesis and cell cycle of the reported betabaculovirus SpobNPV-Manipur isolate. In contrast, the baculovirus lacks the replication-related genes helicase 2, p35, ie-2 and ribonucleotide reductase subunits (rr1 and rr2). The p35 and ie-2 genes were reported to stimulate the DNA replication [47] and subunits of ribonucleotide reductase enzymes catalyze the reduction of host rNTPs to dNTPs and assist in the viral replication [53].
| Gene type | Core genes | Conserved genes in Lepidoptera baculovirus | Other baculovirus genes |
|---|---|---|---|
| SpobNPV | SpobNPV | SpobNPV | |
| Replication | lef2 (SpobNPV8), lef1 (SpobNPV17), DNA-pol (SpobNPV33), helicase (SpobNPV71), alk-exo (SpobNPV140) | ie-1 (SpobNPV25), lef3 (SpobNPV31), lef7 (SpobNPV38), lef11 (SpobNPV57), dbp (SpobNPV93), me53 (SpobNPV143) | ac79 (SpobNPV118), pcna (SpobNPV129) |
| Transcription | vlf-1 (SpobNPV86), p47 (SpobNPV91), lef5 (SpobNPV110), lef4 (SpobNPV113,114), lef9 (SpobNPV120), lef8 (SpobNPV128) | pk-1 (SpobNPV5), 39k (SpobNPV58), lef6 (SpobNPV96) | pe38 (SpobNPV2, 3), lef12 (SpobNPV 90), ie-0 (SpobNPV142) |
| Structure | odv-e18 (SpobNPV21), odv-ec27 (SpobNPV22), desmoplakin (SpobNPV32), ac53 (SpobNPV44,126), vp39 (SpobNPV46) odv-ec43 (SpobNPV51) p48/p45 (SpobNPV63,64), p33 (SpobNPV66, 67), p18 (SpobNPV68), odv-e25 (SpobNPV70), ac81 (SpobNPV83), gp41 (SpobNPV84), ac78 (SpobNPV85), p40 (SpobNPV108), P6.9 (SpobNPV109) 38k (SpobNPV115),vp1054 (SpobNPV125), 49k (SpobNPV141) |
polyhedrin (SpobNPV1), F protein (SpobNPV56), p12 (SpobNPV106), calyx/pep (SpobNPV112) | viral capsid protein (SpobNPV6,7,62), odv-e26 (SpobNPV19), gp64 (SpobNPV35), cg30 (SpobNPV47), odv-e (SpobNPV 69), pkip (SpobNPV92), p24 (SpobNPV134), gp16 (SpobNPV135), p10 (SpobNPV137) |
| Oral infection |
pif5 (SpobNPV26), pif6 (SpobNPV30), pif2 (SpobNPV55), pif4 (SpobNPV72), pif1 (SpobNPV73), pif3 (SpobNPV76), ac110 (SpobNPV82), vp91/p95 (SpobNPV111), p74 (SpobNPV136) | ac108 (SpobNPV50,77,78) | odv-e66 (SpobNPV132) |
| Auxiliary | - | 38.7k (SpobNPV16), ubiquitin (SpobNPV59) | bro-e (SpobNPV11), ptp (SpobNPV13), egt (SpobNPV18), iap-2 (SpobNPV28, 117), MTase (SpobNPV29), gp37 (SpobNPV34), cathepsin (SpobNPV 36), chitinase (SpobNPV 37), arif-1 (SpobNPV54), fgf (SpobNPV61), bro-b protein (SpobNPV65), bro-a (SpobNPV75), iap-1 (SpobNPV 95), ac30 (SpobNPV 98), iap-3 (SpobNPV 101), sod (SpobNPV104), ctl (SpobNPV12, 105), bro-k (SpobNPV116), bro-d (SpobNPV119), bjdp (SpobNPV127), gta (SpobNPV133), p26 (SpobNPV138) |
| Unknown | - | ac145 (SpobNPV23), ac146 (SpobNPV24), ac106 (SpobNPV49), ac76 (SpobNPV87), ac75 (SpobNPV88) | ORF4 peptide (SpobNPV4), ORF146 peptide (SpobNPV9), ac4 (SpobNPV10), ac11 (SpobNPV14,15), ORF140 (SpobNPV16), ac17 (SpobNPV20), Ep Protein (SpobNPV27), ac124 (SpobNPV39), ORF33 peptide (SpobNPV40), ac120 (SpobNPV41), hypothetical protein (SpobNPV48,74), ac18 (SpobNPV52), ac19 (SpobNPV53), ac34 (SpobNPV60), Spob038(SpobNPV79), Spob039 (SpobNPV80), ac111 (SpobNPV81), ac74 (SpobNPV89), ORF113 peptide (SpobNPV94), ac29 (SpobNPV97), Spob106 (SpobNPV99), Spob107 (SpobNPV100), Spob109 (SpobNPV102), ORF121 peptide (SpobNPV103), Spob048 (SpobNPV107), ChaB-like (SpobNPV121), ac59 (SpobNPV122), ac57 (SpobNPV123), ac55 (SpobNPV124), Etm (SpobNPV130), Spob095 (SpobNPV131), Spob021 (SpobNPV139), ORF18 (SpobNPV144) |
Table 5: Gene Content of SpobNPV Manipur isolate
Among the 31 SpobNPV structural genes, 18 genes belong to the baculovirus core genes, 4 genes belong to the lepidopteran conserved genes and 9 genes belong to other baculovirus genes. The polyhedrin/ granulin, polyhedron envelope/calyx, enhancin, p10 and alkaline protease were the key baculovirus genes reported to be associated with baculovirus occlusion bodies [48,54]. Among them, the polyhedrin, polyhedron envelope/calyx and p10 genes were identified in the genome dataset of SpobNPV-Manipur isolate. Certain baculoviruses possess two or three copies of the desmoplakin gene [55]. We have observed only one copy of desmoplakin in the annotated genome of the SpobNPV virus.
The auxiliary genes were not essential for the baculovirus replication and structure but provide a selective advantage for their survival in nature [56]. Among the thoroughly studied auxiliary genes, we identified the lepidopteran conserved genes egt, chitinase and cathepsin [57,58] in the genome dataset of the SpobNPV virus. The egt gene encodes an enzyme, which participates in conjugating the insect molting hormone, ecdysteroid with UDP-glucose [59]. The deletion of the egt gene in baculovirus confirmed that the egt expression is essential for the suppression of insect larval molting [60]. Besides, the cysteine protease genes chitinase and cathepsin facilitate the release of virus occlusion bodies from the insect by breaking down the chitin and the cuticular protein [57]. The deletion of either chitinase or cathepsin can prevent the liquification of the host insect and the host remains intact for several days post their death [61]. In addition, we identified 5 baculovirus repeated ORFs (bro genes) in the SpobNPV genome. The bro genes were found in most of the baculoviruses and possess the ability of DNA binding and nucleosome association to influence the host DNA replication and transcription [62].
Homologous repeat regions (hrs) and bro genes in Spob- NPV genome
Most of the baculovirus genomes contain homologous repeat regions (hrs), characterized by the presence of rich AT content, tandem repeat sequences and imperfect palindromes, interspersed throughout the genome [63,64]. The homologous repeat regions (hrs) are highly variable in nature and exhibit limited homology within different baculoviruses [65]. The tandem repeat finder identified a total of 5 hrs in SpobNPV-Manipur isolate genome dataset (File S2). This is the first report regarding the presence of hrs in the SpobNPV virus as the hrs repeat sequences were missing in the previously reported genome of SpobNPV IIPR (Akram et al., 2018 (Unpublished work)). The size of the SpobNPV hrs direct repeats varied from 35 bp to 99 bp. Besides, the AT content of SpobNPV-Manipur isolates ranged from 53% - 71%.
Simultaneously, we have also identified 5 copies of bro (Baculovirus Repeated Open Reading Frames) genes: bro-a, bro-b, bro-d, bro-e and bro-k in the genome of SpobNPV-Manipur isolate (Table 4). Whereas, the bro-c and bro-b-r genes reported previously in the genome of SpobNPV IIPR (Akram et al., 2018 (Unpublished work)) were missing in the SpobNPV-Manipur isolate.
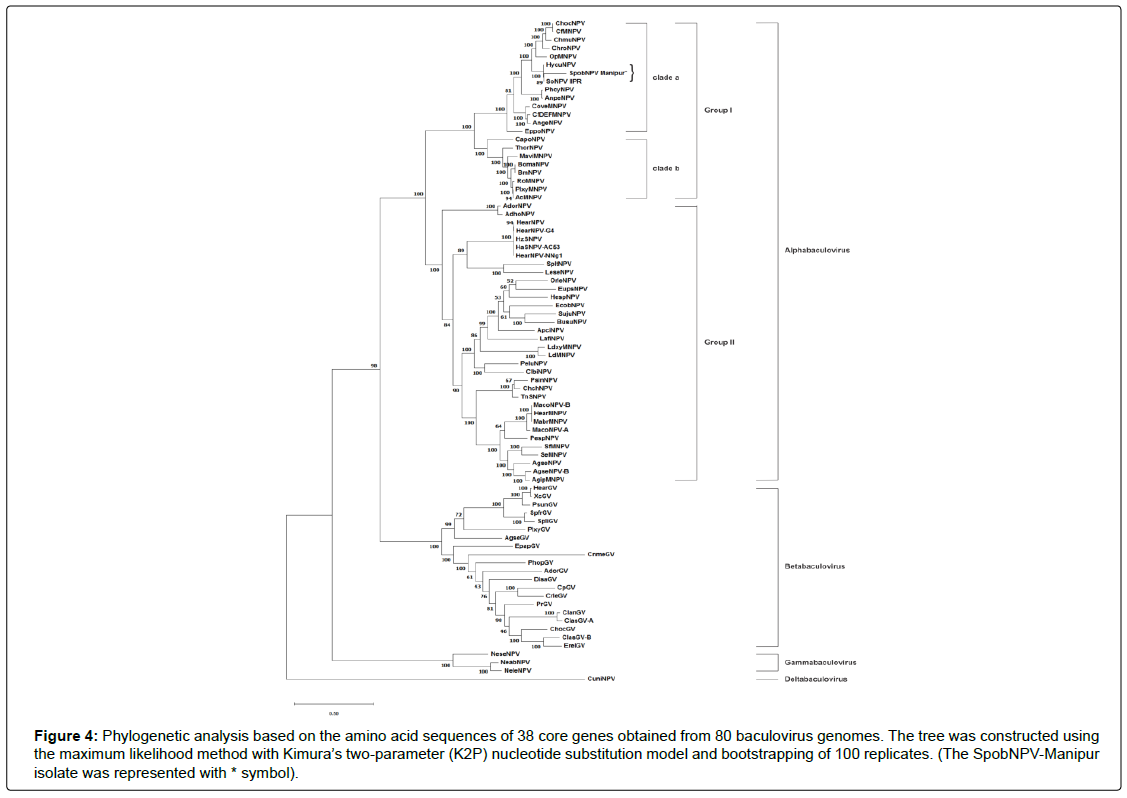
Phylogenetic analysis of SpobNPV core genes
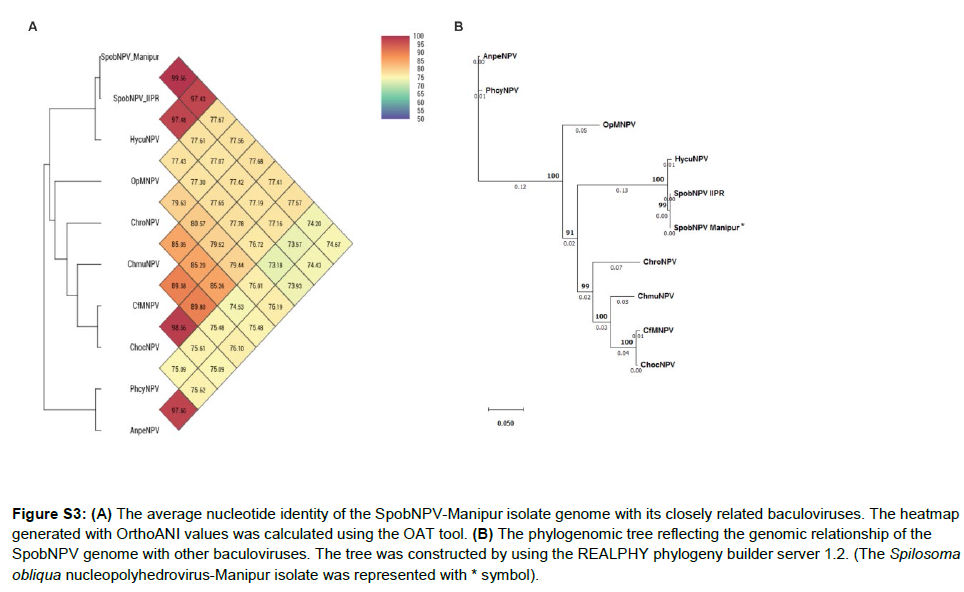
A Phylogenetic tree was constructed based on 38 core genes obtained from a total of 80 baculoviruses (including SpobNPVManipur isolate). The baculovirus replication related genes polyhedrin and DNA polymerase have been used previously for phylogenetic analysis [66]. The per os infectivity factors and transcription specific genes lef8 and lef9 were considered as reliable baculovirus markers for phylogenetic analysis to identify the lepidopteran baculoviruses and monitoring their diversity [67,68]. Besides, the lef2 gene-based phylogeny in HearNPV was demonstrated as a useful parameter to interpret the ancestral relationship and evolutionary history of the baculovirus [69]. The phylogenetic tree represented in our study based on the core genes of the SpobNPV-Manipur isolate provided a clear evolutionary classification between the four genera of the baculovirus (Alphabaculovirus, Betabaculovirus, Gammabaculovirus and Deltabaculovirus) likewise the phylogeny of CapoNPV [65] and CnmeGV [70]. The species of the alphabaculovirus genera were further subdivided into two groups (group I and group II). The phylogenetic analysis placed the SpobNPV-Manipur isolate in clade “a” within the group I alphabaculoviruses (Figure 4). According to the phylogenetic tree, the selected core genes of SpobNPV-Manipur isolate showed close evolutionary relatedness to their orthologs in SpobNPV IIPR (Spilosoma obliqua nucleopolyhedrovirus isolate IIPR) (Akram et al., unpublished work) and HycuNPV (Hyphantria cunea nucleopolyhedrovirus) [71] as they were grouped together as a monophyletic clade and identified as the most recent common ancestors (Figure 4).
Figure 4: Phylogenetic analysis based on the amino acid sequences of 38 core genes obtained from 80 baculovirus genomes. The tree was constructed using the maximum likelihood method with Kimura’s two-arameter (K2P) nucleotide substitution model and bootstrapping of 100 replicates. (The SpobNPV-Manipur isolate was represented with * symbol).
Comparison of SpobNPV ORFs with other baculoviruses
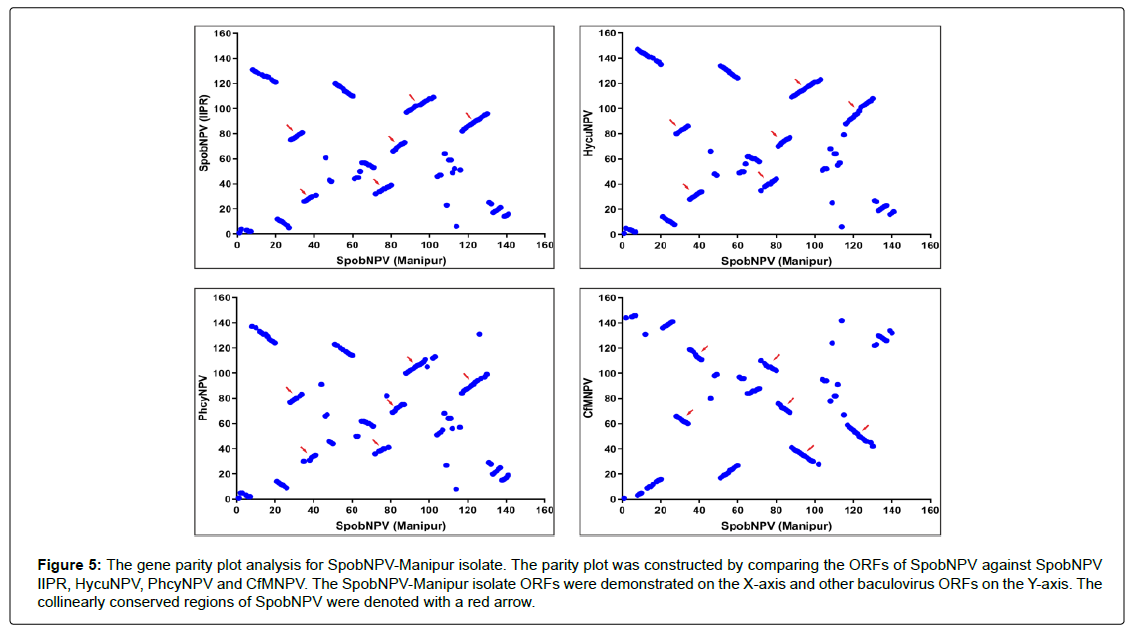
The SpobNPV-Manipur isolate ORFs were compared to their homologues in 4 nucleopolyhedroviruses (SpobNPV IIPR, HycuNPV, PhcyNPV and CfMNPV obtained from the clade “a” of the group I NPV phylogenetic tree). The ORF comparison study demonstrated that SpobNPV-Manipur isolate shared 131, 138, 122 and 127 homologous ORFs with SpobNPV IIPR, HycuNPV, PhcyNPV and CfMNPV respectively (Table 4). The ORFs sequence similarity analysis denoted that the SpobNPV-Manipur isolate ORFs exhibited an average amino acid (aa) identity of 98%, 97%, 80% and 84% with their homologous ORFs in the selected 4 group I NPVs. For the core genes, SpobNPV showed average aa identity of 98%, 98%, 86% and 89% with its baculoviruses homologues. Besides, the SpobNPV exhibited 98%, 97%, 81% and 84% average aa identity for the lepidopteran conserved genes and 99%, 97%, 77% and 81% identity for the other baculovirus genes with their homologous ORFs in the selected NPVs. A total of 20 SpobNPV-Manipur isolate ORFs were identified with sequence identity of 90% and above against their homologues in all the four selected baculoviruses.
The gene parity plot assists in comparing the position of the orthologous genes (gene orders) in different genomes and assessing the synteny conservation within the genomes [66]. The gene parity plot analysis of SpobNPV-Manipur isolate (Figure 5) with the selected baculoviruses obtained from the same clade demonstrated moderate co-linearity with inverted regions over the whole genome likewise the parity plot of BusuNPV [32] and contrary to the plots of HycuGVCpGV and HycuGV- PlxyGV [72]. In contrast to the parity plot distribution of most of the baculoviruses [31,73,72] the gene parity plot of SpobNPV-Manipur isolate showed the presence of multiple collinearly conserved regions identified between SpobNPV28 and SpobNPV34, SpobNPV35 and SpobNPV41, SpobNPV73 and SpobNPV81, SpobNPV83 and SpobNPV89, SpobNPV90 and SpobNPV104 and SpobNPV120 and SpobNPV133. In SpobNPVManipur isolate the collinearly conserved regions contain 12 core genes, 6 lepidopteran conserved genes and 31 other baculovirus genes.
Figure 5: The gene parity plot analysis for SpobNPV-Manipur isolate. The parity plot was constructed by comparing the ORFs of SpobNPV against SpobNPV IIPR, HycuNPV, PhcyNPV and CfMNPV. The SpobNPV-Manipur isolate ORFs were demonstrated on the X-axis and other baculovirus ORFs on the Y-axis. The collinearly conserved regions of SpobNPV were denoted with a red arrow.
Multi genome comparison and phylogenomic relationship with other baculoviruses
The CLC assembled 41 SpobNPV genomic contigs were concatenated using Geneious Prime sequence analysis software version 2019.1. The concatenated genome dataset of SpobNPV-Manipur isolate was aligned against Antheraea pernyi nucleopolyhedrovirus (AnpeNPV) (NC_008035) [43], Choristoneura fumiferana multiple nucleopolyhedrovirus (CfMNPV) (NC_004778) [73], Choristoneura murinana nucleopolyhedrovirus (ChmuNPV) (NC_023177) [74] and Spilosoma obliqua nucleopolyhedrovirus isolate IIPR (SpobNPV IIPR) (KY550224) (Akram et al., (Unpublished work)) group I alphabaculoviruses genomes reported in NCBI using the Mauve multiple genome alignment tool. The Mauve genome alignment generated 28, 28, 30, 29, 29, 35, 28, 28 and 38 Locally Collinear Blocks (LCBs) between the genomes of SpobNPV-Manipur and AnpeNPV, SpobNPV-Manipur and CfMNPV, SpobNPV-Manipur and ChmuNPV, SpobNPV-Manipur and ChocNPV, SpobNPV-Manipur and ChroNPV, SpobNPV-Manipur and HycuNPV, SpobNPVManipur and OpMNPV, SpobNPV-Manipur and PhcyNPV and SpobNPV-Manipur and SpobNPV-IIPR with minimum LCB weight of 75, 146, 128, 125, 130, 146, 201, 80 and 198 respectively (Figure 6).
The concatenated genome dataset of SpobNPV-Manipur isolate exhibited Average Nucleotide Identity (ANI) score [75] of 74.67%, 77.41%, 77.68%, 77.57%, 77.56%, 97.43%, 77.67%, 74.20% and 99.56% with AnpeNPV, CfMNPV, ChmuNPV, ChocNPV, ChroNPV, HycuNPV, OpMNPV, PhcyNPV and SpobNPV IIPR genomes (Figure S3A). The high ANI scores between the SpobNPVManipur and SpobNPV-IIPR and SpobNPV-Manipur and HycuNPV were well supported by their phylogenetic relationships based on the core genes (Figure 4). The phylogenomic analysis based on the genome sequence comparison of SpobNPV-Manipur isolate virus species with their nearest baculoviruses was performed by using the REALPHY phylogeny builder web tool. The phylogenomic tree of the SpobNPV-Manipur isolate denoted close evolutionary relationship of the virus with SpobNPV IIPR and HycuNPV as they were grouped as a monophyletic clade (Figure S3B). The pathogenicity and structural information of the SpobNPV-Manipur isolate virus can be utilized to improve its efficacy in pest management and the genome dataset can be used as a valuable resource to interpret its genetic and molecular mechanisms and promote the virus as an effective bioinsecticide.
Conflict of Interests
The authors declare no potential conflicts of interest.
Acknowledgements
AA thanks Department of Biotechnology, India for SRF (DBT/2015/ MSU/447) and all authors thank DBT Bioinformatics Infrastructure Facility (BT/ BI/04/055/2001) at Manonmaniam Sundaranar University for providing instrument facilities. Bioassay and augmentation of the virus were standardized during the project sponsored by the DBT for which RV would like to thank DBT, New Delhi.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- Yengkhom RS. Biology and control of Bihar hairy caterpillar Spilosoma obliqua walker artiidae lepidoptera.
- Rajen Singh Y (2005) Infestation level of Spilarctia obliqua (Arctiidae: Lepidoptera) on certain crops in Imphal valley of Manipur. Indian J Agri Sci 75(1): 61-62.
- Singh Y, Varatharajan R (2000) Certain aspects of NPV infecting the larvae of Spilosoma obliqua Walker (Arctiidae). Microbials in Insect Pest Management Oxford and IBH Publishing Co Pvt Ltd, New Delhi 133-140.
- Singh OP (1989) Efficacy of some insecticides against eggs and larvae of bihar hairy caterpillar, Spilosoma obliqua Walker infesting mustard crop Brassica campestris. Plant Protection Bulletin Faridabad 41 (1-2): 45-48.
- Singh Y, Varatharajan R (2005) Infestation level of Spilarctia obliqua (Arctiidae: Lepidoptera) on certain crops in Imphal valley of Manipur. Indian J Agri Sci 75 (1): 61-62.
- Hunter-Fujita FR, Entwistle PF, Evans HF, Crook NE (1998) Insect viruses and pest management. John Wiley & Sons Ltd.
- Smith KM (1976) Virus-insect relationships.
- Jacob A, Thomas M (1972) Nuclear-polyhedrosis virus of Diacrisia obliqua (Wlk)(Arctiidae, Lepidoptera). Agric Res J Kerala 10(2).
- Jacob A, Thomas M (1975) Nature of inclusion bodies of a nuclear polyhedrosis virus of Diacrisia obliqua (Walker). Agri Res J Kerala 12 (1): 82-93.
- Chaudhari S (1997) Effect of age of Spilosoma obliqua larvae on their susceptibility to nuclear polyhedrosis virus. Indian J Entomol 59(1): 59-61.
- Battu G, Ramakrishnan N, Prakash N (1991) Size and Shape of Inclusion Bodies of Nuclear Polyhedrosis Virus of Spilosoma obliqua (Walker). J Biol Control 5(2): 88-92.
- Manickavasagam S, Ramakrishnan N, Anuradha S, Prasad Y (1992) Identification of three nuclear polyhedroviruses through restriction endonuclease analysis. J Biol Control 6(2): 101-103.
- Varatharajan R, Singh M, Tandon P, Ballal C, Jallali (2002) In vivo production of Spilarctia obliqua (Walker) NPV. In: S, Rabindra R (eds) Symposiumof Biological Control of Lepidopteran Pests, Bengaluru Society for Biocontrol Advancement Pp: 149-152.
- Kumar CS, Jacob T, Devasahayam S, D’Silva S, Jinsha J, Rajna S, et al. (2015) Occurrence and characterization of a tetrahedral nucleopolyhedrovirus from Spilarctia obliqua (Walker). J Invertebr Pathol 132: 135-141.
- Sudhakar S, Mathavan S (1999) Electron microscopical studies and restriction analysis ofHelicoverpa armigera nucleo polyhedrosis virus. J Biosci 24 (3): 361-370.
- Sudhakar S, Varatharajan R, Mathavan S (1997) Simple method to purify polyhedral inclusion bodies from Nosema (Microspora: Nosematidae) contamination. ENTOMON-TRIVANDRUM 22: 89-94.
- Rabindra R, Rajasekaran B, Jayaraj S (1997) Combined action of nuclear polyhedrosis virus and neem bitter against Spodoptera litura (Fabricius) larvae. J Biol Control 11: 5-9.
- Abbott W (1925) A method of computing the effectiveness of an insecticide. J Econ Entomol 18: 265-267.
- Finney D (1977) Probit analysis. Cambridge University Press, London.
- Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53(282): 457-481.
- Sudhakar S, Varatharajan R, Mathavan S (1997) Simple method to purify polyhedral inclusion bodies from Nosema (Microspora: Nosematidae) contamination. Entomon 22: 89-94.
- O'Reilly DR, Miller LK, Luckow VA (1994) Baculovirus expression vectors: a laboratory manual. Oxford University Press on Demand.
- Andrews S (2016) FastQC: a quality control tool for high throughput sequence data 2010.
- Gallardo-Escárate C, Valenzuela-Muñoz V, Nuñez-Acuña G (2014) RNA-Seq analysis using de novo transcriptome assembly as a reference for the salmon louse Caligus rogercresseyi. PloS one 9(4): e92239.
- IJkel WF, Van Strien EA, Heldens JG, Broer R, Zuidema D, et al. (1999) Sequence and organization of the Spodoptera exigua multicapsid nucleopolyhedrovirus genome. J Gen Vir 80(12): 3289-3304.
- Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, et al. (2008) NCBI BLAST: a better web interface. Nucleic acids research 36(suppl_2): W5-W9.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28 (12): 1647-1649.
- 28. Alikhan NF, Petty NK, Zakour NLB, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12(1): 402.
- Rohrmann G (2008) Baculovirus Molecular Biology. Bethesda (MD): National Library of Medicine (US). National Center for Biotechnology Information Pp: 1-154.
- Garcia-Maruniak A, Maruniak JE, Zanotto PM, Doumbouya AE, Liu J-C, et al. (2004) Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J Vir 78(13): 7036-7051.
- . Alikhan NF, Petty NK, Zakour NLB, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12(1): 402.
- Zhu SY, Yi JP, Shen WD, Wang LQ, He HG, et al. (2009) Genomic sequence, organization and characteristics of a new nucleopolyhedrovirus isolated from Clanis bilineata larva. BMC Genomics 10 (1): 91.
- Zhu Z, Yin F, Liu X, Hou D, Wang J, et al. (2014) Genome sequence and analysis of Buzura suppressaria nucleopolyhedrovirus: a group II Alphabaculovirus. PLoS One 9 (1): e86450.
- Jehle JA, Blissard G, Bonning B, Cory J, Herniou E, et al. (2006) On the classification and nomenclature of baculoviruses: a proposal for revision. Arch Virol 151(7): 1257-1266.
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31(13): 3497-3500.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Bio Evol 33(7): 1870-1874.
- Wennmann JT, Keilwagen J, Jehle JA (2018) Baculovirus Kimura two-parameter species demarcation criterion is confirmed by the distances of 38 core gene nucleotide sequences. J Gen Vir 99 (9): 1307-1320.
- Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14(7): 1394-1403.
- Lee I, Kim YO, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2): 1100-1103.
- Devi WS, Varatharajan R Granulosis Virus of the cabbage pest, pieris brassicae–a potential biopesticide for the control of pieris spp.(pieridae: lepidoptera).
- https://doi.org/10.1093/molbev/msu088
- Xu Y-P, Ye Z-P, Niu C-Y, Bao Y-Y, Wang W-B, et al. (2010) Comparative analysis of the genomes of Bombyx mandarina and Bombyx mori nucleopolyhedroviruses. J Microbiol 48(1): 102-110.
- Gomi S, Majima K, Maeda S (1999) Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J Gen Vir 80 (5): 1323-1337.
- Nie Z-M, Zhang Z-F, Wang D, He P-A, Jiang C-Y, et al. (2007) Complete sequence and organization of Antheraea pernyi nucleopolyhedrovirus, a dr-rich baculovirus. BMC Genomics 8(1): 248
- Vlak JM, Smith GE (1982) Orientation of the genome of Autographa californica nuclear polyhedrosis virus: a proposal. J Virology 41(3): 1118-1121.
- Xing K, Deng R, Wang J, Feng J, Huang M, et al. (2005) Analysis and prediction of baculovirus promoter sequences. Virus Res 113(1): 64-71.
- Vanarsdall AL, Mikhailov VS, Rohrmann GF (2007) Baculovirus DNA replication and processing. Curr Drug Targets 8(10): 1096-1102.
- Rohrmann GF (2008) Baculovirus molecular biology.
- Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM (1994) Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proceedings of the National Academy of Sciences 91(23): 11212-11216.
- Fu Y, Wang R, Liang A (2018) Function analysis of Ac-PCNA and Sf-PCNA during the Autographa californica multiple nucleopolyhedrovirus infection process. Mol Cell Biochem 443(1-2): 57-68.
- Crawford AM, Miller LK (1988) Characterization of an early gene accelerating expression of late genes of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virology 62(8): 2773-2781.
- Lu A, Miller LK (1995) The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virology 69(2): 975-982.
- Mitchell JK, Byers NM, Friesen PD (2013) Baculovirus F-box protein LEF-7 modifies the host DNA damage response to enhance virus multiplication. J Virology 87(23): 12592-12599.
- Lange M, Jehle JA (2003) The genome of the Cryptophlebia leucotreta granulovirus. Virology 317(2): 220-236
- Blissard GW, Theilmann DA (2018) Baculovirus entry and egress from insect cells. Annual review of virology 5(1): 113-139.
- O’Reilly DR (1997) Auxiliary genes of baculoviruses. In: The baculoviruses. Springer, 267-300.
- Wang J, Hou D, Wang Q, Kuang W, Zhang L, et al. (2018) Genome analysis of a novel Group I alphabaculovirus obtained from Oxyplax ochracea. PLoS One 13(2): e0192279.
- Cory J (2001) Host manipulation by insect pathogens: the effect of the baculovirus egt gene on the host-virus interaction. Endocrine interactions of insect parasites and pathogens.
- Cory JS, Myers JH (2003) The ecology and evolution of insect baculoviruses. Annual Review of Ecology, Evolution, and Systematics 34 (1): 239-272.
- Kelly TJ, Park E, Masler C, Burand JP (1995) Characterization of the glycosylated ecdysteroids in the hemolymph of baculovirus-infected gypsy moth larvae and cells in culture. Eur J Entomol 92: 51-51.
- O'Reilly DR, Miller LK (1989) A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245(4922): 1110-1112.
- Hawtin RE, Zarkowska T, Arnold K, Thomas CJ, Gooday GW, et al. (1997) Liquefaction ofAutographa californicaNucleopolyhedrovirus-Infected Insects Is Dependent on the Integrity of Virus-Encoded Chitinase and Cathepsin Genes. Virology 238(2): 243-253.
- Zemskov EA, Kang W, Maeda S (2000) Evidence for nucleic acid binding ability and nucleosome association of Bombyx mori nucleopolyhedrovirus BRO proteins. J Virology 74(15): 6784-6789.
- Kool M, Voeten J, Goldbach R, Tramper J, Vlak J (1993) Identification of seven putative origins of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus DNA replication. J Gen Virol 74(12): 2661-2668.
- Hilton S, Winstanley D (2008) The origins of replication of granuloviruses. Arch Virol 153 (8): 1527-1535.
- Wang J, Zhu Z, Zhang L, Hou D, Wang M, et al. (2016) Genome sequencing and analysis of Catopsilia pomona nucleopolyhedrovirus: a distinct species in group I alphabaculovirus. PLoS One 11(5): e0155134.
- Herniou EA, Olszewski JA, Cory JS, O'Reilly DR (2003) The genome sequence and evolution of baculoviruses. Annu Rev Entomol 48(1):211-234.
- Herniou EA, Olszewski JA, O'reilly DR, Cory JS (2004) Ancient coevolution of baculoviruses and their insect hosts. J Virology 78(7): 3244-3251.
- Jehle JA, Lange M, Wang H, Hu Z, Wang Y, et al. (2006) Molecular identification and phylogenetic analysis of baculoviruses from Lepidoptera. Virology 346(1): 180-193.
- Chen X, IJkel WF, Dominy C, de Andrade Zanotto PM, Hashimoto Y, et al. (1999) Identification, sequence analysis and phylogeny of the lef-2 gene of Helicoverpa armigera single-nucleocapsid baculovirus. Virus Res 65(1): 21-32.
- Han G, Xu J, Liu Q, Li C, Xu H, et al. (2016) Genome of Cnaphalocrocis medinalis Granulovirus, the First Crambidae-Infecting Betabaculovirus Isolated from Rice Leaffolder to Sequenced. PLoS One 11(2): e0147882.
- Ikeda M, Shikata M, Shirata N, Chaeychomsri S, Kobayashi M (2006) Gene organization and complete sequence of the Hyphantria cunea nucleopolyhedrovirus genome. J Gen Virol 87(9): 2549-2562.
- De Jong JG, Lauzon HA, Dominy C, Poloumienko A, Carstens EB, et al. (2005) Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. J Gen Virol 86(4): 929-943.
- Rohrmann GF, Erlandson MA, Theilmann DA (2014) Genome sequence of an alphabaculovirus isolated from Choristoneura murinana. Genome Announc 2(1): e01135-01113.
- Arahal DR (2014) Whole-genome analyses: average nucleotide identity. In: Methods in microbiology, vol 41: 103-122.
| Summary | SpobNPV |
|---|---|
| Total reference length (bp) | 136,306 |
| Number of mapped reads | 508,000 |
| mapped read length (bp) | 58,085,035 |
| Maximum coverage | 5,996 |
| Average coverage | 424.65 |
| Standard deviation | 226.60 |
Table S1: Genome coverage statistics of SpobNPV Manipur isolate.
| Variant type | Variant subtype | # variants |
|---|---|---|
| Deletion | Self mapped | 19 |
| Deletion | Paired breakpoint | 2 |
| Deletion | Cross mapped breakpoints | 7 |
| Total (Deletion) | 28 | |
| Insertion | Self mapped | 6 |
| Insertion | Paired breakpoint | 1 |
| Insertion | Close breakpoints | 103 |
| Insertion | Tandem duplication | 4 |
| Total (Insertion) | 114 | |
| Inversion | Cross mapped breakpoints | 9 |
| Inversion | Paired breakpoint | 0 |
| Total (Inversion) | 9 | |
| Replacement | Paired breakpoint | 20 |
| Total (Replacement) | 20 | |
| Translocation | Multiple breakpoints | 0 |
| Total (Translocation) | 0 | |
| Complex | Can't resolve sequence | 0 |
| Complex | Multiple breakpoints | 16 |
| Complex | Cross mapped breakpoints (invalid orientation) | 3 |
| Total (Complex) | 19 | |
| Total (InDels and Structural Variants) | 190 |
Table S2: Summary of InDels and Structural Variants identified between the Spilosoma obliqua NPV (KY550224.1) and Spilosoma obliqua NPVManipur isolate genomes.
| Reference genome Position | Type | Spilosoma obliqua NPV IIPR Nucleotide (Reference) | Spilosoma obliqua NPV Manipur isolate Nucleotide (Test) |
|---|---|---|---|
| 8744..8745 | MNV | GG | AA |
| 27984..27985 | MNV | CA | AT |
| 35689..35690 | MNV | GC | AT |
| 53302..53303 | MNV | CC | TT |
| 53351..53352 | MNV | AA | GC |
| 54314..54315 | MNV | AA | GC |
| 54827..54828 | MNV | GG | AA |
| 54882..54883 | MNV | GG | AA |
| 55321..55323 | MNV | ATT | CGC |
| 55340..55341 | MNV | TT | CC |
| 55667..55668 | MNV | CG | TA |
| 81708..81709 | MNV | TT | CC |
| 85078..85079 | MNV | TT | CC |
| 134659..134660 | MNV | TG | CC |
| 1707 | SNV | G | A |
| 1741 | SNV | G | A |
| 3516 | SNV | G | A |
| 3752 | SNV | A | G |
| 4269 | SNV | G | A |
| 4530 | SNV | T | C |
| 5239 | SNV | C | T |
| 5323 | SNV | G | A |
| 5426 | SNV | A | G |
| 5680 | SNV | A | G |
| 7029 | SNV | A | G |
| 7228 | SNV | G | A |
| 8413 | SNV | C | T |
| 8514 | SNV | C | T |
| 8799 | SNV | A | G |
| 8811 | SNV | G | T |
| 8815 | SNV | C | T |
| 8821 | SNV | C | T |
| 8942 | SNV | A | G |
| 9001 | SNV | C | T |
| 9050 | SNV | A | T |
| 9185 | SNV | T | G |
| 9359 | SNV | G | A |
| 9361 | SNV | G | C |
| 9380 | SNV | T | A |
| 9396 | SNV | T | A |
| 10508 | SNV | A | G |
| 10750 | SNV | A | T |
| 11508 | SNV | T | C |
| 12186 | SNV | C | T |
| 12378 | SNV | C | T |
| 13773 | SNV | G | A |
| 15382 | SNV | T | A |
| 16764 | SNV | G | A |
| 16904 | SNV | G | A |
| 16911 | SNV | T | C |
| 17125 | SNV | A | G |
| 17150 | SNV | G | A |
| 17438 | SNV | C | T |
| 19693 | SNV | C | A |
| 19742 | SNV | G | A |
| 19931 | SNV | T | C |
| 20009 | SNV | T | G |
| 20173 | SNV | G | A |
| 20175 | SNV | C | G |
| 21106 | SNV | G | A |
| 23217 | SNV | C | T |
| 26053 | SNV | C | T |
| 26230 | SNV | C | T |
| 26644 | SNV | A | G |
| 26749 | SNV | C | T |
| 27885 | SNV | T | G |
| 27994 | SNV | G | A |
| 28116 | SNV | T | C |
| 30447 | SNV | A | G |
| 30573 | SNV | T | C |
| 32347 | SNV | T | C |
| 33431 | SNV | C | T |
| 33715 | SNV | T | C |
| 34925 | SNV | A | G |
| 34934 | SNV | A | C |
| 34945 | SNV | G | A |
| 34948 | SNV | C | T |
| 35410 | SNV | C | T |
| 35545 | SNV | T | C |
| 35547 | SNV | C | T |
| 35559 | SNV | A | G |
| 35574 | SNV | G | A |
| 35607 | SNV | G | A |
| 35678 | SNV | T | C |
| 35687 | SNV | A | G |
| 35815 | SNV | T | C |
| 35827 | SNV | A | G |
| 35875 | SNV | G | A |
| 35942 | SNV | G | A |
| 35994 | SNV | T | A |
| 36229 | SNV | A | G |
| 36261 | SNV | A | G |
| 36877 | SNV | G | A |
| 38325 | SNV | G | A |
| 40094 | SNV | A | C |
| 40155 | SNV | T | C |
| 40317 | SNV | T | C |
| 40494 | SNV | G | A |
| 40554 | SNV | C | T |
| 40587 | SNV | A | G |
| 40728 | SNV | A | G |
| 40731 | SNV | G | T |
| 41521 | SNV | T | G |
| 41590 | SNV | G | A |
| 41602 | SNV | T | C |
| 42542 | SNV | C | A |
| 43021 | SNV | T | C |
| 44337 | SNV | C | T |
| 45620 | SNV | C | G |
| 45683 | SNV | T | C |
| 45839 | SNV | C | G |
| 46081 | SNV | C | T |
| 46151 | SNV | T | C |
| 46204 | SNV | C | T |
| 47233 | SNV | C | T |
| 48723 | SNV | G | A |
| 49143 | SNV | G | C |
| 52702 | SNV | A | G |
| 52872 | SNV | A | G |
| 53103 | SNV | A | C |
| 53109 | SNV | G | A |
| 53163 | SNV | T | C |
| 53202 | SNV | A | G |
| 53217 | SNV | A | G |
| 53223 | SNV | A | G |
| 53244 | SNV | A | G |
| 53250 | SNV | G | T |
| 53343 | SNV | G | C |
| 53349 | SNV | T | C |
| 53355 | SNV | T | C |
| 53361 | SNV | G | A |
| 53403 | SNV | A | C |
| 53445 | SNV | T | G |
| 53451 | SNV | G | A |
| 53454 | SNV | C | T |
| 53456 | SNV | C | T |
| 53466 | SNV | T | C |
| 53472 | SNV | G | A |
| 53504 | SNV | T | C |
| 54024 | SNV | C | T |
| 54273 | SNV | A | C |
| 54306 | SNV | G | C |
| 54312 | SNV | T | C |
| 54318 | SNV | T | C |
| 54321 | SNV | A | C |
| 54324 | SNV | T | A |
| 54657 | SNV | G | A |
| 54693 | SNV | C | A |
| 54700 | SNV | C | A |
| 54710 | SNV | T | C |
| 54713 | SNV | G | A |
| 54745 | SNV | T | G |
| 54783 | SNV | T | C |
| 54786 | SNV | G | A |
| 54837 | SNV | T | C |
| 54861 | SNV | G | C |
| 54904 | SNV | T | C |
| 55164 | SNV | G | C |
| 55221 | SNV | C | T |
| 55263 | SNV | A | G |
| 55281 | SNV | C | T |
| 55299 | SNV | G | A |
| 55311 | SNV | G | C |
| 55319 | SNV | C | T |
| 55326 | SNV | A | C |
| 55347 | SNV | T | C |
| 55410 | SNV | C | G |
| 55535 | SNV | A | G |
| 55541 | SNV | G | A |
| 55570 | SNV | G | T |
| 55590 | SNV | A | T |
| 55671 | SNV | C | T |
| 55683 | SNV | A | G |
| 55687 | SNV | C | T |
| 55740 | SNV | T | C |
| 56250 | SNV | C | T |
| 56982 | SNV | T | C |
| 57962 | SNV | C | T |
| 60046 | SNV | A | G |
| 60802 | SNV | A | G |
| 61159 | SNV | A | G |
| 61372 | SNV | T | C |
| 62122 | SNV | C | T |
| 62146 | SNV | C | T |
| 62152 | SNV | G | A |
| 62170 | SNV | C | T |
| 62176 | SNV | G | A |
| 62183 | SNV | C | G |
| 62185 | SNV | A | G |
| 62191 | SNV | G | A |
| 63547 | SNV | C | T |
| 64486 | SNV | A | G |
| 65576 | SNV | A | G |
| 65580 | SNV | T | C |
| 65714 | SNV | G | A |
| 65872 | SNV | T | G |
| 66766 | SNV | C | G |
| 67271 | SNV | T | C |
| 69267 | SNV | G | A |
| 74847 | SNV | T | C |
| 76632 | SNV | C | T |
| 76727 | SNV | G | T |
| 77109 | SNV | T | G |
| 77778 | SNV | T | C |
| 78034 | SNV | T | C |
| 81272 | SNV | G | T |
| 81432 | SNV | C | A |
| 84067 | SNV | A | G |
| 84099 | SNV | A | G |
| 84587 | SNV | A | G |
| 84747 | SNV | C | G |
| 85784 | SNV | G | A |
| 86168 | SNV | A | G |
| 86252 | SNV | C | T |
| 88313 | SNV | G | A |
| 88350 | SNV | C | T |
| 90370 | SNV | T | C |
| 93557 | SNV | T | C |
| 95216 | SNV | A | G |
| 95813 | SNV | G | A |
| 95907 | SNV | T | G |
| 95909 | SNV | G | C |
| 96055 | SNV | C | G |
| 96576 | SNV | T | C |
| 101739 | SNV | G | A |
| 102833 | SNV | C | T |
| 103567 | SNV | C | T |
| 103752 | SNV | G | A |
| 103773 | SNV | C | T |
| 104280 | SNV | A | G |
| 104862 | SNV | G | T |
| 105176 | SNV | T | G |
| 107389 | SNV | G | A |
| 107451 | SNV | A | G |
| 107677 | SNV | T | C |
| 108549 | SNV | G | A |
| 108573 | SNV | C | T |
| 108844 | SNV | G | A |
| 108867 | SNV | A | G |
| 110407 | SNV | G | A |
| 111585 | SNV | A | G |
| 112020 | SNV | G | T |
| 112279 | SNV | G | C |
| 112377 | SNV | C | T |
| 112681 | SNV | C | T |
| 112687 | SNV | C | T |
| 112967 | SNV | A | T |
| 112991 | SNV | T | C |
| 112997 | SNV | T | G |
| 113086 | SNV | C | T |
| 113431 | SNV | C | T |
| 113507 | SNV | G | A |
| 113906 | SNV | G | A |
| 114660 | SNV | G | A |
| 115549 | SNV | C | T |
| 116863 | SNV | A | G |
| 121098 | SNV | T | C |
| 121296 | SNV | C | T |
| 122108 | SNV | C | T |
| 122682 | SNV | T | C |
| 123229 | SNV | T | G |
| 123737 | SNV | C | T |
| 125244 | SNV | T | C |
| 125960 | SNV | G | C |
| 130002 | SNV | G | A |
| 130404 | SNV | G | A |
| 130503 | SNV | G | A |
| 131289 | SNV | A | G |
| 131372 | SNV | A | C |
| 132600 | SNV | C | T |
| 133526 | SNV | A | G |
| 133728 | SNV | G | A |
| 133877 | SNV | G | A |
| 134631 | SNV | C | G |
| 135655 | SNV | C | T |
Table S3: List of Variants identified between the SpobNPV IIPR and SpobNPV Manipur isolate genomes.
| Reference genome length (bp) | 136,141 |
| GC contents in reference genome (%) | 45.50 |
| Number of mapped reads | 252,930 |
| Length of mapped reads (bp) | 30,046,318 |
| Maximum coverage | 649 |
| Average coverage | 219.25 |
| Size of assembled genome (bp) | 136020 |
| GC% of assembled genome | 45.40 |
File S1: Summary statistics of reference-based assembly in SpobNPV-Manipur isolate.
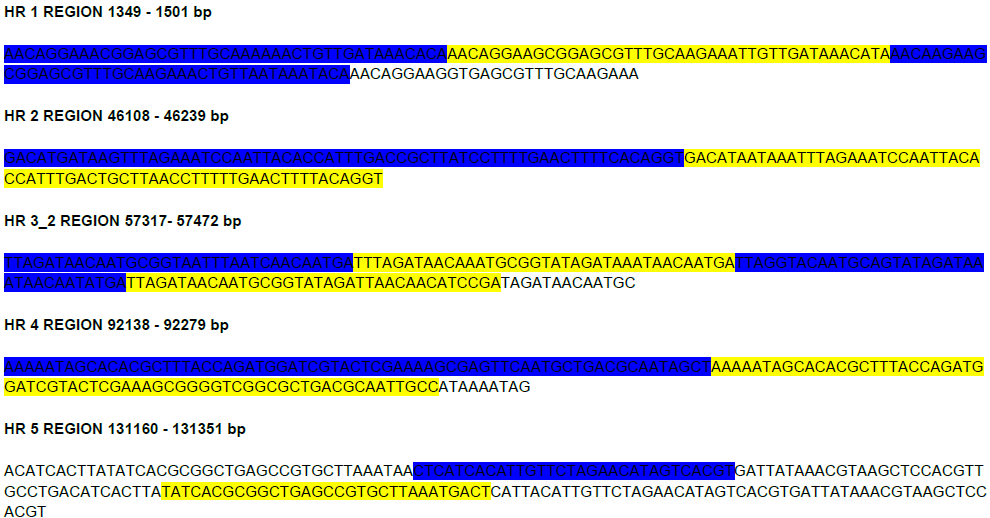
File S2: Alignment of SpobNPV-Manipur homologous repeats (hrs).
Figure S1: Statistics of sequencing read quality. Histogram representing the (A) length and (B) Phred quality score distribution of the raw reads (after trimming) in the SpobNPV-Manipur isolate genome. (C) Histogram representing the length distribution of de novo assembled contigs in the SpobNPV virus. (D) The coverage distribution of the raw reads in the assembled genome of SpobNPV-Manipur isolate.
Figure S3: (A) The average nucleotide identity of the SpobNPV-Manipur isolate genome with its closely related baculoviruses. The heatmap generated with OrthoANI values was calculated using the OAT tool. (B) The phylogenomic tree reflecting the genomic relationship of the SpobNPV genome with other baculoviruses. The tree was constructed by using the REALPHY phylogeny builder server 1.2. (The Spilosoma obliqua nucleopolyhedrovirus-Manipur isolate was represented with * symbol).
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi