Research Article, J Appl Bioinforma Comput Biol Vol: 6 Issue: 3
In Silico Inhibition Studies of AXL Kinase by Curcumin and its Natural Derivatives
Ghrifi Fatima*, Allam Loubna, Lakhlili Wiame and Ibrahimi Azeddine
The Biotechnology Lab (MedBiotech), Rabat Medical and Pharmacy School, Mohammed V University in Rabat, Morocco
*Corresponding Author : Ghrifi Fatima
Biotechnology Lab (MedBiotech), Rabat Medical and Pharmacy School, Mohammed V University in Rabat, Avenue Mes belarbi Alaoui, Suissi-Rabat, BP6203 Rabat instituts, Rabat,10000, Morocco
Tel: +212 537 77 28 50
E-mail: ghrifatima1000@gmail.coms
Received: August 08, 2017 Accepted: November 07, 2017 Published: November 13, 2017
Citation: Fatima G, Loubna A, Wiame L, Azeddine I (2017) In Silico Inhibition Studies of AXL Kinase by Curcumin and its Natural Derivatives. J Appl Bioinforma Comput Biol 6:3. doi: 10.4172/2329-9533.1000142
Abstract
The last few years have seen the intensive investigation efforts for understanding the wide relevance of AXL activation in multiple aspects of oncogenesis, invasion, metastasis and drug resistance. Therfore, targeting AXL can be considered as one of the promissing approaches for the treatement of cancer. A series of curcumin derivatives which are natural polyphenolic coumponds were wellreported as an anti cancer and chemopreventive agents. We have investigated them as an ATP-competitive inhibitors of AXL kinase by using a docking studies. We built a model of AXL catalytic domain from the crystal structure of proto-oncogene tyrosine-protein kinase MER (MERTK) and the modeling of the three-dimensional (3D) structure of the AXL was performed by SWISS-MODEL homology modeling program. The quality and validation of the model were performed using PROCHECK and VERIFY 3D softwares. The Ramachandran plot was used to validate the overall stereochemical property of the protein. All curcuminoids formed a hydrogen bond with hinge region by Methionine (Met 623) and lysine (Lys 567) indicating that these compounds can be utilized therapeutically as a natural AXL inhibitors.
Keywords: AXL kinase; Curcumin; Docking; Homology modeling
Introduction
The AXL receptor tyrosine kinase , also known as UFO, Tyro7 and Ark, is a member of the tyrosine kinases receptors family TAM (Tyro3, AXL and MERTK) [1]. AXL, like the other TAM members, is activated via the interaction with the growth arrest-specific protein 6 (GAS6) ligand [2]. The receptor has been implicated in a number of oncogenic processes. AXL signalization enhances many essential biological functions for cancer formation and progression, including invasion, migration, survival, angiogenesis, cell transformation and proliferation [3]. The increase of AXL activity or overexpression indicates a poor prognosis for the patients and has been reported to be associated with metastasis in several types of cancer [4-6]. AXL was demonstrated to confer resistance to chemotherapies when overexpressed in gastrointestinal stromal tumor [7] and breast cancer cells [8]. AXL was first cloned from myeloid leukemia cells in 1991[9].No crystal structure for Axl catalytic domain has been reported. Just the extracellular Ig-like domain was crystallized as fragments in complex with Gas6 in 2006 [10].
During the last decades, inhibition of AXL tyrosine kinases has emerged as an important approach for cancer therapy. Until this date, no inhibitor was clearly designed and marketed to inhibit AXL. However, Some clinical molecules, developed to inhibit other kinases, were evaluated against AXL given its interest, but these molecules continue to be inadequate having either limited efficacy, prohibitive toxicities or often exhibit less potency for AXL. All these are due to the similarity of the kinases catalytic domains, to which classical ATP competitive inhibitors (type I and II) tend to bind, particularly, with c-MET and MERTK kinases. Recently, the first AXL specific small molecule inhibitor, BGB324 originally discovered by BergenBio Company as R-428, entered phase 1 clinical trials [11].
Thus, there is a need for other therapies that can desactivate AXL kinase and keep the cancer in remission and increase survival. Previous studies have focused on the anticancer effects of natural products, for designing novel molecules ATP competitives with low toxicity.
Curcumin, has been shown to be effective against different cancers [12-14] in both in vitro and in vivo assays through a variety of mechanisms [15,16]. In addition, human clinical trials did not demonstrate any toxicity for curcumin at doses up to 10 g/day [17]. Curcumin, isolated from turmeric Curcuma longa contains curcumin as major component(77%) but it also contains demethoxycurcumin (DMC) (17%) and bisdemethoxycurcumin (BDMC) (3%) that have a remarkable anti- carcinogenisis effect. A study showed that demethoxycurcumin induced the G2/M step of cellular cycle arrest in human glioma U87cells [18]. In another study, BDMC was observed to suppress the growth and activity in human adenogastric carcinoma [19].
Here, we focused on examining the interactions between curcumin as well as DMC and BDMC with AXL domain kinase using in silico appraoch and molecular docking. This study could be applied to develop new potential AXL inhibitors ATP competitives from curcumin and natural derivatives in the future. Since no data are available on AXL crystal structure. Herein we used homology modeling to this end.
Materials and Methods
AXL kinase homology modeling
We used the Swiss model server [20] to bluit the AXL model by homology modeling, a proto-oncogene tyrosine-protein kinase MER (MERTK PDB code:3BRB, resolution 1.9Å) was used as template.
The template was selected based on sequence identity and the coverage of the sequence obtained by alignement using Basic Local Alignment SearchTool [21] NCBI BLAST. AXL and MERTK sequences were obtained from UniProt database (ID: P30530; ID: Q12866 respectively) Uniprot.
Model validation
Structural refinement was further assessed by Structural Analysis and Verification Server (version3) (nih server) This meta server runs six programs for checking and validating protein structures. Verify3D analyzes the compatibility of an atomic model (3D) with its own amino acid sequence (1D) [22]. The structure was analyzed by Ramachandran’s plot using PROCHECK software [23].
Active site validation
In order to evaluate the model catalytic cavity and critical residues involved in AXL active site, molecular docking studies of ATP with the AXL model was performed with AutoDock vina 4.0 [24]. The best conformation with best binding affinity in kcal/mol was selected.
Docking approach
Binding mode and selectivity of AXL kinase with curcumin and naturals derivatives were studied by AutoDock vina 4.0 [24] which required the ligand and receptor in pdbqt format and the configuration file as txt format. The Autodock Vina also produces files containing the ligands docked pose as output wich contain the score of the binding affinity, these files were analyzed to study interactions and binding energy of the docking. The results were visualized using Pymol molecular viewer v.1.7.4 schrodinger.
Ligands collection and preparation
Curcumin and two natural analogues namly demetoxycurcumin (DMC), bisdemethoxycurcumin (BDMC) coumponds were selected on the basis of their anticarcinogenic activity experimentally proved and metabolic stability in comparaison with native curcumin. The strucutres of these coumponds were downloaded from NCBI Pubchem (CID:96516; 5469424; 5315472 respectively) in PDB format. The Pymol was used to verify the presence or absence of hydrogen, the stereochemistry of chiral carbons, number of rotable bonds, number of hydrogen acceptors and donors.
For all ligands, Gasteiger charges were designated and the torsions for ligands were permitted to rotate during docking procedure using Autodock tools v.1.5.6. The ligands were saved in pdbqt format.
Target preparation
The receptor was prepared in Autodock tools v.1.5.6, polar hydrogens were assigned to the receptor, the grid-box was centered around the AXL active site, the dimensions were set at 30,30,30 Å (x, y and z) consisting of active site residues within the box. The receptor was saved in pdbqt format.
Result
Sequence analysis
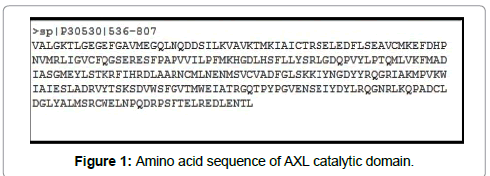
The amino acid sequence of AXL catalytic domain was retrieved from the Uniprot database. The selected sequence for physiochemical analysis and 3D modeling is given in Figure 1.
Sequence alignment by BLAST showed, 70% identity between a sequence of the AXL catalytic domain and the template.
3D structure prediction and model validation
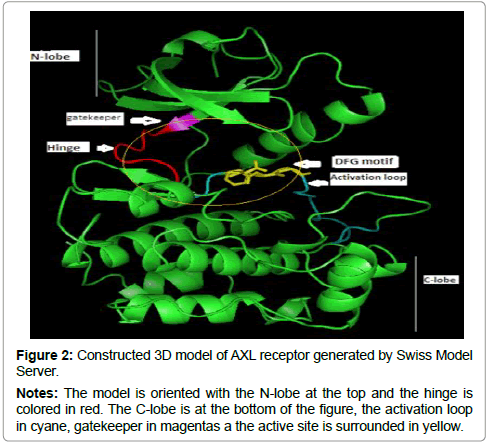
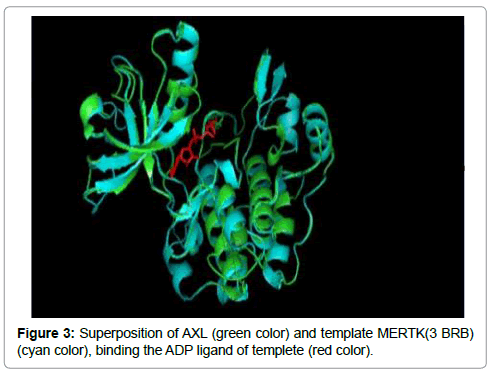
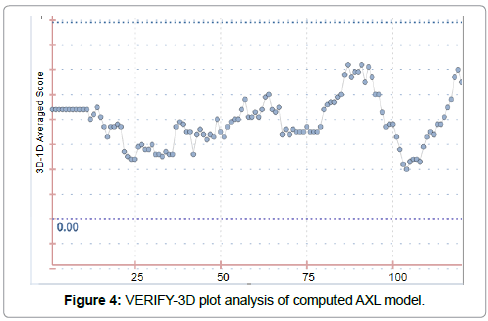
The 3D structure prediction (Figure 2) was carried out by alignment of target sequences with template structure (PDB ID 3BRB), using a Swiss Model Server. Figure 3 presents the supperposition between the constructed model and the template complexed with ADP ligand. Figure 4 shows the VERIFY-3D graph determining that 95.96% of AXL residues have an average score of 3D-1D ≥ 0.2 confirming the compatibility of the 3D structure and the primary sequence of AXL.
Figure 2: Constructed 3D model of AXL receptor generated by Swiss Model Server.
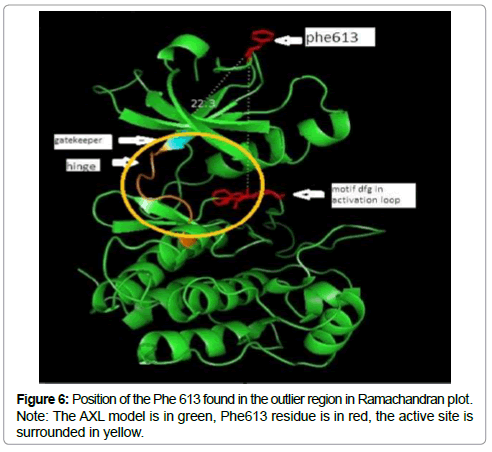
Notes: The model is oriented with the N-lobe at the top and the hinge is colored in red. The C-lobe is at the bottom of the figure, the activation loop in cyane, gatekeeper in magentas a the active site is surrounded in yellow.
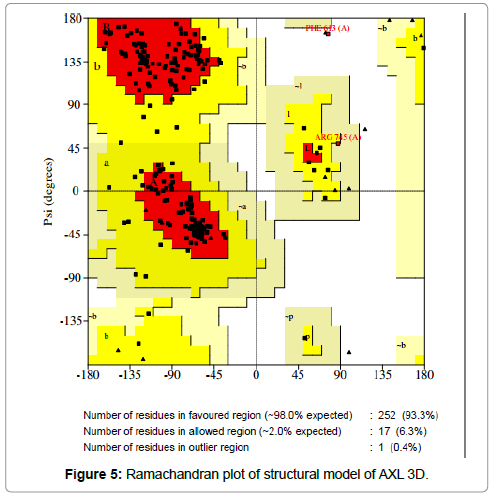
The sterochemical quality and accuracy of the predicted model was evaluated after the refinement process using the Ramachandran plot calculated with the PROCHECK program [23]. The π and ψ distributions of the Ramachandran plots of non-glycine, non-proline residues are represented in Figure 5, showing 93.3% (252 aminoacids) of residues in the favoured regions, 6.3% (17amino acids) in allowed region and 0.4% (1 amino acids) in outlier region Figure 6 shows the residue phenylalanine (Phe613) found in the outlier region is at very long distances from the pocket suggesting that this amino acid is structurally and functionally irrelevant to the ligand-binding site.
Active site validation by ATP
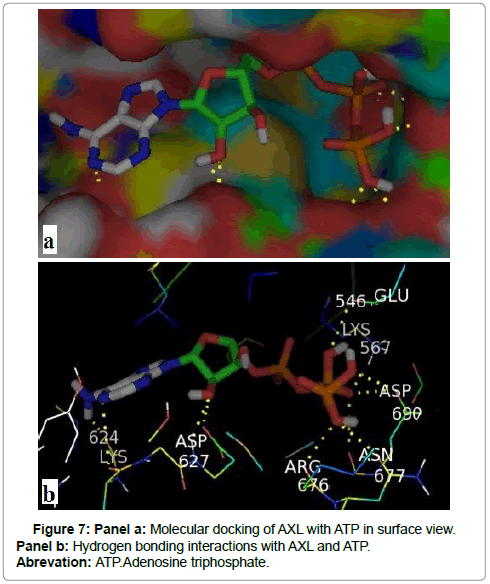
Among the nine conformations results of docking between AXL kinase and ATP, we have selected the best binding position with an energy of -8.7 kcal /mol. Figure 7 shows that AXL kinase interacts with ATP principally by seven hydrogen bonds which are summarized in Table 1.
| Residues | H Bond length (Å) |
|---|---|
| Glu 546 Lys 567 Lys 624 Asp 627 Arg676 Asn677 Asp690 |
3.0 1.9 2.1 2.4 3.3 2.1 3.3 |
Table 1: Molecular docking results of AXL receptor with ATP using Vina docking software.
Interaction betwenn AXL and curcumin compounds
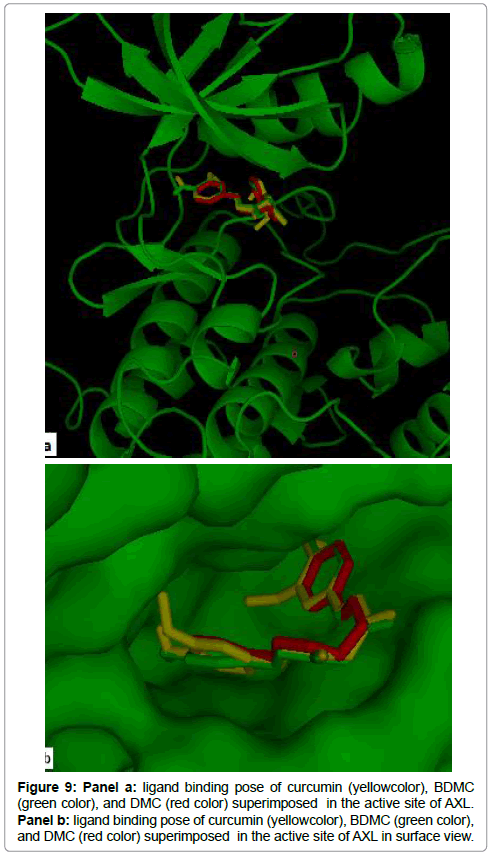
The 2D structures of curcumin compounds used in this study are illustrated in Figure 8. As showed in Figure 9(a,b), the three compounds occupied the common binding site of the AXL kinase. The binding energy of AXL- curcumin, AXL-DMC and AXL-BDMC complexes was respectivelly -9.8 kcal/mol, -9.4 kcal/mol and -9.0 kcal/ mol. Table 2 ressemble the binding affinity and the hydrogen bond distances of the key residues that interact with inhibitors.
| Ligands docked With AXL |
binding affinity Kcal /mol |
Interaction amino acid Residues | H Bond distance (Å) |
|---|---|---|---|
| Curcumin | -9.8 | Lys567 Met623 Ala689 Asp690 |
2.27 2.30 2.50 3.122 |
| Demetoxycurcumin (DMC) | -9.4 | Lys 567 Met623 Ala689 Asp690 |
1.3 2.7 3.2 3.4 |
| Bisdemethoxy curcumin (BDMC) |
-9.0 | Lys 567 Met 623 Ala 689 Asp690 |
2.20 2.40 3.26 3.06 |
Table 2: The binding affinity and the hydrogen bond distances of the key residues that interact with inhibitors.
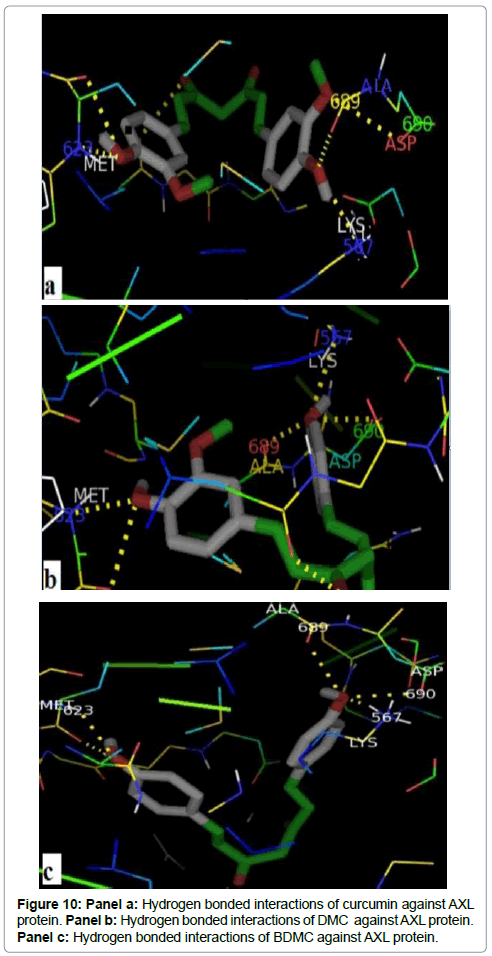
Figures 10 (a, b and c) shows that all curcuminoids formed a hydrogen bonds with the same residues, Met 623, Lys 567, Ala 689 and Asp 690. The Met 623 of the hinge region makes a hydrogen bonds of 2.3Å, 2.7Å and 2.4Å between their nitrogen backbone with curcumin, DMC and BDMC respectivelly. The Lys 567, one of the binding site key residues of ATP, interacts with curcuminoid compounds, the shorter one have a distance of 1.3 Å in their interaction with DMC. The shorter distance between Ala 689 of the activation loop and curcumin coumpond was 2.5 Å. Furthermore, the three compounds possess a hydrogen bond with DFG motif by their Asparatic acid residue. The hydrogen bonds were created between the side chain of Asp690 and curcumin, DMC and BDMC have a distance of 3.12Å, 3.4Å and 3.06Å respectively.
Discussion
In this paper, we used the in Silico approach and molecular docking to explain the binding mechanism of curcumin and its natural derivatives (DMC, BDMC) to the AXL kinase. We generated a model of AXL kinase using the MERTK as a template for the Swiss Model software and the model was evaluated by Verrify 3D and PROCHECK programs. The model validation indicated the acceptable quality of the structural model with 99.6% of the model’s torsion angles in favorable positions. The molecular docking did confirm that the ATP molecule is positioned in their AXL kinase binding site. Thereafter, we studied the curcuminoids interaction with the AXL kinase to demonstrate if these molecules hold a position inside of the ATP binding site, the results obtained from the molecular docking demonstrated that the selected phytoconstituents are positioned into the active cavity of AXL and exhibits good binding mode with AXL.
The binding affinity of the curcuminoids with AXL is well above that of ATP. The curcuminoids create hydrogen bonds with three of the key residues in ATP binding site. The first one was with Met 623 of the hinge region; the second was with Lys 567 positioned on a β-sheet of N-lobe, and the third with Asp 690 of the DFG-motif. Curcumin was found to bind with the best affinity with AXL than DMC and BMDC. The difference in affinity may be due to the number of methoxy groups on the aromatic rings in the curcuminoids, both natural derivatives are two congeners of curcumin isolated from turmeric, furthermore curcumin has two symmetric phenyl-methoxy groups, DMC contains one and BMDC contains none. This suggests that the phenyl methoxy groups do contribute to the inhibiting power of compounds Chen and al demonstrated that the activity is also due to the ortho-methoxy phenolic functionality [25].
Curcumin, together with demethoxycurcumin and bisdemethoxycurcumin, are the three predominant active compounds derived from the turmeric root. Several reports have shown that curcumin could modulate each stage of cancer such as initiation, promotion and progression [26] Kim et al. indicate that curcumin has inhibitory effects on AXL expression as Gas6-dependent AXL phosphorylation, and AXL promoter activity in non-small cell lung cancer cells [27]. The present results will be the basis to develop new compounds ATP- competitive more selective and active against AXL kinase, derived from the curcumin molecule. These results encouraged us to perform the ongoing structural-activity relationship studies to evaluate the relationship of the methoxy-groups and the inhibitory effect against AXL.
Conclusion
Traditionally, the turmeric has been used commonly as a spice in curries to give a specific flavor and yellow color. This phytochimical and it derivatives have also been found to be effective against different cancers. Our results showed that these compounds have a strong binding affinity to the AXL receptor and could be useful as a source of potential therapeutics or lead molecules for prevention or treatment of multiple cancers. Curcumin has a promising cytotoxic effects against cancer cell lines. But, further clinical research about the toxicity of the molecule is needed to bring in more clarity on human safety and how their activity could be exercised in vivo after human consumption.
Competing Interests
The authors declare that they have no competing interests.
Aknowledgments
This work was carried out under National funding from the Moroccan Ministry of Higher education & Scientific research (PPR program) to AI. This work was also supported by a grant from the NIH for H3Africa BioNet to AI.
References
- Lemke G (2013) Biology of the TAM receptors. Cold Spring Harb Perspect Biol 5: a009076.
- Mark MR, Chen J, Hammonds RG, Sadick M, Godowsk PJ (1996) Characterization of Gas6, a member of the superfamily of G domain-containing proteins, as a ligand for Rse and Axl. J Biol Chem. 271: 9785-9789.
- Linger RMA, Keating AK, Earp HS, Graham DK (2008) TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res 100: 35-83.
- Shieh YS, Lai CY, Kao YR, Shiah SG, Lee HS, et al. (2005) Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia 7: 1058-1064.
- Wu CW, Li AFY, Chi CW, Lai CH, Huang CL, et al. (2002) Clinical significance of AXL kinase family in gastric cancer. Anticancer Res 22: 1071-1078.
- Sainaghi PP, Castello L, Bergamasco L, Galletti M, Bellosta P, Avanzi GC (2005) Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J Cell Physiol 204: 36-44.
- Mahadevan D, Cooke L, Riley C, Swart R, Simons B, et al. (2007) A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene 26: 3909-3919.
- Li Y, Ye X, Tan C, Hongo JA, Zha J, et al. (2009) Axl as a potential therapeutic target in cancer: role of Axl in tumor growth, metastasis and angiogenesis. Oncogene 28: 3442-3455.
- O’Bryan JP, Frye RA, Cogswell PC, Neubauer A, Kitch B, et al. (1991) axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol 11: 5016-5031.
- Sasaki T, Knyazev PG, Clout NJ, Cheburkin Y, Göhring W, et al. (2006) Structural basis for Gas6-Axl signalling. EMBO J 25: 80-87.
- Sheridan C (2013) First Axl inhibitor enters clinical trials. Nat Biotechnol 31: 775-776.
- Huang MT, Smart RC, Wong CQ, Conney AH (1988) Inhibitory effect of curcumin, chlorogenic acid, caffeic acid, and ferulic acid on tumor promotion in mouse skin by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res 48: 5941-5946.
- Kuttan G, Kumar KBH, Guruvayoorappan C, Kuttan R (2007) Antitumor, anti-invasion, and antimetastatic effects of curcumin. Adv Exp Med Biol 595: 173-184.
- Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, et al. (2007) Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol 72: 29-39.
- Kunnumakkara AB, Anand P, Aggarwal BB (2008) Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett 269:199-225.
- Thangapazham RL, Sharma A, Maheshwari RK (2006) Multiple molecular targets in cancer chemoprevention by curcumin. AAPS J 8:E443-449.
- Aggarwal BB, Kumar A, Bharti AC (2003) Anticancer potential of curcumin: preclinical and clinical studies. Anticancer Res 23: 363-398.
- Luthra PM, Kumar R, Prakash A (2009) Demethoxycurcumin induces Bcl-2 mediated G2/M arrest and apoptosis in human glioma U87 cells. Biochem Biophys Res Commun 384: 420-425.
- CL, ZD, XW, GC, ZF (2015) Bisdemethoxycurcumin attenuates gastric adenocarcinoma growth by inducing mitochondrial dysfunction. Oncol Lett 9: 270-274.
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinforma Oxf Engl 22: 195-201.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403-410.
- Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277: 396-404.
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477-486.
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31: 455-461.
- Chen WF, Deng SL, Zhou B, Yang L, Liu ZL (2006) Curcumin and its analogues as potent inhibitors of low density lipoprotein oxidation: H-atom abstraction from the phenolic groups and possible involvement of the 4-hydroxy-3-methoxyphenyl groups. Free Radic Biol Med 40: 526-535.
- Patel VB, Misra S, Patel BB, Majumdar APN (2010) Colorectal cancer: chemopreventive role of curcumin and resveratrol. Nutr Cancer 62: 958-967.
- Kim KC, Baek SH, Lee C (2015) Curcumin-induced downregulation of Axl receptor tyrosine kinase inhibits cell proliferation and circumvents chemoresistance in non-small lung cancer cells. Int J Oncol 47: 2296-2303.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi