Research Article, J Appl Bioinforma Comput Biol Vol: 11 Issue: 7
In-silico Evaluation of the Genes and Proteins Involved in Bacterial Bio-plastic Production Pathways
Hamza Abbas Jaffari, Qamar Abbas* and Hafiz Zeshan Wadood
Department of Biology, Lahore Garrison University, Lahore, Pakistan
*Corresponding Author: Qamar Abbas
Department of Biology,
Lahore Garrison
University,
Lahore,
Pakistan,
Tel: 923349422223;
E-mail: qamar92@lgu.edu.pk
Received date: 20 May, 2022, Manuscript No. JABCB-22-64407; Editor assigned date: 23 May, 2022, PreQC No. JABCB-22-64407 (PQ); Reviewed date: 06 June, 2022, QC No. JABCB-22-64407; Revised date: 19 July, 2022, Manuscript No. JABCB-22-64407 (R); Published date: 26 July, 2022, DOI: 10.4172/2329-9533.1000251.
Citation: Jaffari HA, Abbas Q, Wadood HZ (2022) In-silico Evaluation of the Genes and Proteins Involved in Bacterial Bio-plastic Production Pathways. J Appl Bioinforma Comput Biol 11:7.
Abstract
Plastic pollution is a worldwide concern endangering both fossil fuel resources and the environment through rise in their production and usage. Bioplastics emerged as a suitable replacement and proved more environment friendly than traditional plastics. Among bioplastics, Polyhydroxyalkanoates (PHA) proved to be in demand due to their promising properties such as high rate of biodegradability. Three key genes (phaA, phaB, and phaC ) are involved in the biosynthesis of PHA. An analysis based on bioinformatics approaches was performed in this study to analyze these key genes and their translated proteins in potential PHA producing microorganisms found on the gen-solution database. The sequences (DNA and Protein) obtained from PHA producing microorganisms were analyzed in MEGA X to check their evolutionary history. The arrangement of genes was highly diverse. The PHA biosynthetic genes belonging to Proteobacteria showed promising phylogenetic relationship and the occurrence and their distribution in these organisms suggested that a single gene cluster related to PHA might have been segmented in the form of small groups of genes and got transferred among various organisms. The results indicated that the gene and protein sequences were not fully conserved suggesting that the genes had undergone addition, deletion or replacement of nucleotides resulting in gene rearrangement. However, the function of the proteins remained fully conserved suggesting that the protein function had the highest level of conservation and the gene sequences had the least level of conservation according to this study. Overall, the results were a strong indicative that horizontal gene transfer had a strong influence on the distribution and molecular evolution of these PHA biosynthetic genes.
Keywords: Polyhydroxyalkanoates; phaA, phaB; phaC; Bioplastics; PHA synthase
Introduction
Petroleum based plastics have become a necessity and their production has increased dramatically over the past 75 years. Plastic usage is expected to be doubled in the next 20 years [1]. Synthetic plastics are among the most environmentally harmful substances produced by mankind i.e., occupying the near top position [2].
Because conventional plastics are cheap, versatile, persistent in the environment and have a poor biodegradation rate, their improper disposal has become a global issue [3,4]. A lot of countries have developed special programs toward the discovery of new substances new strategies intended at changing pollution to make it easier.
Bioplastics are a special biomaterial made from different microbes, which are cultured under different nutritional and environmental conditions [5]. The number and size of granules, monounsaturated compounds, macromolecular structure and physicochemical properties vary, depending on the organism that is producing them [6-9].
Bacterial cells such as Bacillus megaterium, Alcaligenes latus, Cupriavidus necator, along with other archaea and cyanobacteria that synthesize energy or carbon storage materials in the form of hydroxyalkanoate polyesters. Polyhydroxyalkanoates (PHAs) are under consideration as one of the best alternate options with regards to bioplastic production. PHA has up to 150 diverse monomeric structures [10]. Melting temperature, degree of structural order, polymer hydrophobicity and temperature of glass transition totally depends upon the composition of monomers. [11,12].
PHAs are ideal storage compounds because they exert negligible increase in osmotic pressure due to their insolubility in the bacterial cytoplasm [13]. The PHA’s composition and molecular weight is determined by the bacterium type and the conditions required for growth [14].
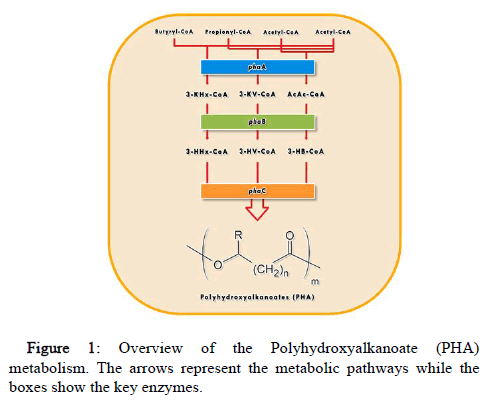
The synthesis of Poly (3-hydroxybutyrate) (PHB) is considered to be one of the first and best PHA biosynthetic pathway representative to be characterized. It involves three steps catalyzed by acetyl-CoA acetyltransferase (β-ketothiolase; phaA), acetoacetyl-CoA reductase (phaB), and PHA synthase (phaC) [15,16].
Various microorganisms have shown this biosynthetic pathway and it has been extensively studied but due to less knowledge of the structural information on the enzymes involved in PHA biosynthesis, the detailed molecular mechanisms involved in the PHA biosynthesis are not that much clear [17,18].
PHAs are intracellular biopolymers that are of uncommon significance due to their physicochemical, biodegradable, and biocorrespondence properties. These materials can be obtained directly from renewable resources, bioplastics which are petroleum based and biologically produced by microorganisms [19]. They can be used for production of food utensils, hygiene items, cosmetic items, glasses, razors, medical surgical clothing, bags, disposable lids and packaging (Figure 1) [20-22].
There are many challenges regarding production of PHAs which are affecting their commercialization as well. One of them is production cost. The synthetic plastics proved to be much cheaper than PHAs in terms of production costs. Efforts have been made to reduce the production cost by selection of cheap and efficient carbon substrates for the production of PHAs [23].
Materials and Methods
Data collection and filtering
There are three main genes involved in the biosynthesis of PHAs i.e., phaA, phaB and phaC. On gen-solution there is an entirely dedicated database for PHA producing organisms obtained by the phylogenetic and statistical analysis of these 3 genes. Gen-solution database provides a lot of relevant and specific information regarding PHA producing microorganisms along with their gene and protein information. The database provided a vast dataset of 23 taxons, 233 organisms and 381 (phaA-111, phaB-199, phaC-71) genes [24].
All the data obtained from this PHA database was downloaded in software compatible format and relevant information was stored in Microsoft Excel.
In data filtering and refining, all collected data was grouped according to the specific genotype i.e., phaA, phaB and phaC respectively. All sequences belonging to eukaryotic taxa were removed to obtain pure prokaryotic results. The sequence data for all phaA, phaB and phaC was normalized on the basis of relative size range.
Later, the size of the gene sequences was observed for a common range. For phaA the range was about 1100-1200 nt, for phaB gene size was ~700-800 nt and for phaC it was approximately 1600-1900 nt. All the sequences that fall in these relative ranges of size were then analyzed.
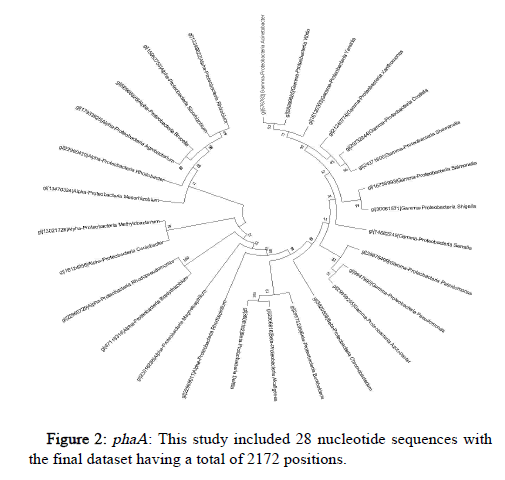
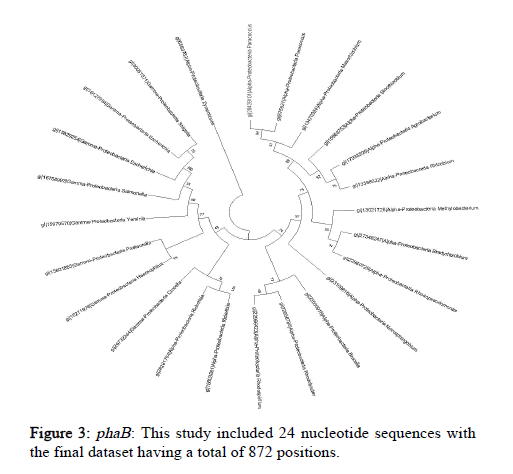
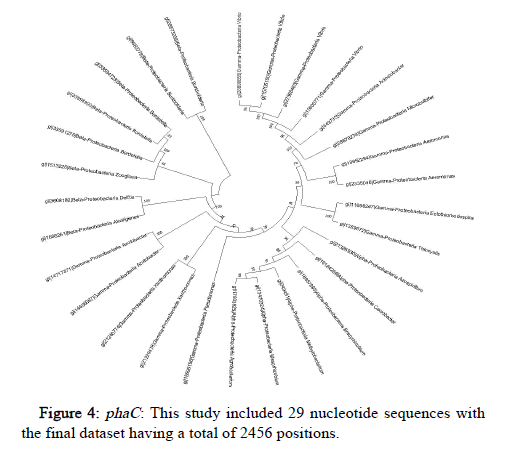
As a result of the data filtering and refining, microorganisms belonging to Proteobacteria were found to be most dominant and were chosen to be analyzed separately.
After analyzing the results of all microorganisms belonging to Proteobacteria, alignment files were generated for each genotype (phaA, phaB, phaC) belonging to each taxon separately i.e., Alpha, Beta, and Gamma.
Sequence alignment using MEGA X
Molecular Evolutionary Genetic Analysis (MEGA) was used for the in-silico analysis of the gene and protein sequences as it provides more efficient algorithms for fast and robust analysis [25].
Sequence data was retrieved and refined for sequence alignment. Sequence alignment was performed to find regions of similarity between sequences that may be due to evolutionary relationships. Individual alignment files were generated for the phaA, phaB and phaC genes, after every phase of data filtering, containing their respective gene sequences in FASTA format with respect to their genotype.
The alignment files of phaA gene, phaB gene and phaC gene were aligned independently using MUSCLE method.
Phylogenetic estimation using MEGA X
There are many widely used methods through which the phylogenetic trees are created and analyzed, such as UPGMA, Maximum Likelihood (ML), Neighbor Joining, Bayesian Inference and Maximum Parsimony but in this research, we used the Maximum Likelihood method.
The bootstrap values were set to 1000 replicates. After completion of the process the constructed phylogenetic tree was displayed.
Protein analysis
Protein sequence retrieval was done along with the gene sequences and the same steps were followed to generate alignment files as for the gene sequences. Data filtering of protein sequences was also done along with the gene sequences. The process of sequence alignment and phylogenetic tree generation was same as explained above for the gene sequences.
Results
Variation of gene sequences in PHA producing microorganisms
Examining the gene and protein sequences after initial dataset filtering and refinement, the results showed that the relationships between the genes, proteins and their phylogeny varies extremely across the tree of life in prokaryotes. Due to these distinct differences in the gene organization, the importance in terms of evolution of these genes and clustering them is underlined and is hence, correlated with the concept of evolution [26]. Moreover, intra-specie sequence data exhibited diversity among the PHA producing microorganisms.
Results showed that the gene sequences are diversely spread among microorganisms producing PHA. This can be due to gene size being altered by different factors such as nucleotide substitution, gene rearrangements due to presence of transposons, integrases and recombinases near the gene cluster of PHA biosynthesis. Genes and protein sequences were then filtered according to a preferred relative size range to avoid diverse results and false positives due to variation in gene sizes. The results obtained showed that conserved regions are not there but there are conserved nucleotides which indicate towards conservation of function even if the sequences are not fully conserved.
Insights in molecular evolution of PHA Biosynthetic genes of Proteobacteria
Analyzing results of initial data filtering and relative size range, it was found that the presence of identical gene clusters is in microorganisms that are closely related particularly belonging to Proteobacteria. But there are also many instances where the location of these gene clusters contains only a subset of these genes and may be interrupted by genes not involved in PHA biosynthesis. A reason for these genes to be different and not contain conserved regions can be due to different consequences such as insertions or deletions in the genetic sequence that may have caused these gene segments to expand elongate and generate a new genetic sequence for the gene in different microorganisms. This suggests that these genes have been linked with horizontal gene transfer and have been shaped extensively by this [27,28].
The Tamura-Nei model and the Maximum Likelihood approach were used here to determine the phylogeny of phaA, phaB and phaC genes of microorganisms belonging to Proteobacteria, represented by a consensus tree generated from 1000 replicates of bootstrap values. MEGA X was used to undertake evolutionary analysis (Figures 2-4).
Phylogenetic analysis of PHA biosynthetic genes in alpha, beta and gamma Proteobacteria
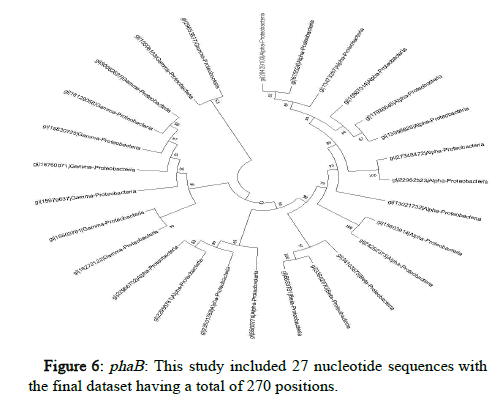
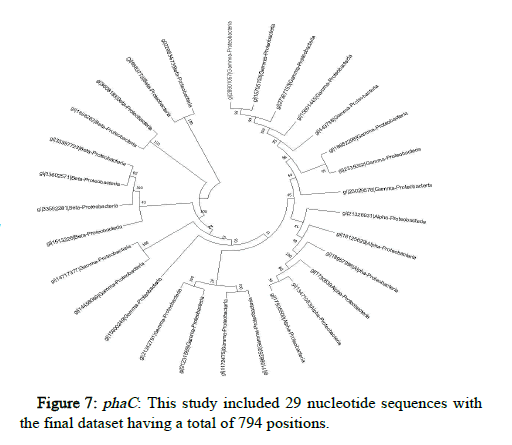
Conserved gene pattern was found in Proteobacteria, particularly alpha, beta and gamma Proteobacteria. The gene clusters appeared to be both common and conserved in these. This investigation focused primarily on determining the phylogenetic relationship of these PHA producers but there was no intention to study how these PHA gene clusters were produced or how they got distributed among different organisms. But still, it has been demonstrated in previously conducted studies that genes that are involved in the metabolism of PHA had experienced independent evolution and have a greater degree of difference as compared to the other genes [28]. The results displayed that majority of the PHA biosynthetic genes were found in the microorganisms belong to the group of alpha, beta and gamma Proteobacteria.
Comparative phylogenetic analysis of proteins involved in PHA biosynthesis
After examining the results of protein sequence analysis and comparing them with the results of gene sequence analysis, it was evident that the conservation of protein sequences was higher than that of the genes. The alignment sessions performed after all phases of data filtration showed that there is higher degree of conservation among protein sequences in comparison to gene sequences.
The JTT matrix-based and the maximum likelihood approach were used here to determine the phylogeny of phaA, phaB and phaC proteins of microorganisms belonging to Proteobacteria, represented by a consensus tree generated from 1000 replicates of bootstrap values. MEGA X was used to undertake evolutionary analysis (Figures 5-7).
Discussion
The PHA pathway is important for free-living strain's environmental fitness, as evidenced by the extensive maintaining of genes clustered in microorganisms living in a variety of different habitats [29,30]. No significant differences in gene content were found in Proteobacteria studied; this suggests that environment is a better and greater factor in determining the PHA gene clusters than the phylogenetic origin. Furthermore, we believe that genes and their clustering make functionally related metabolic genes to shift by Horizontal Gene Transfer (HGT) easier for organisms.
Phylogenetic studies show that once acquired, PHA genes have never been lost during the course of evolution. Clearly, the stepwise and progressive novel PHA gene addition and disruption has caused the cluster size to increase. The clusters are made up of orthologous genes found in closely related or unrelated species.
The order of PHA genes and their neighbors proved to be both diverse and conserved. In closely related bacterial species of Proteobacteria, adjacent genes were kept near to PHA associated gene clusters. This diversification in the rearrangements strongly suggest that genome relocation events had a substantial role in the maintaining the biosynthetic genes i.e., phaA, phaB and phaC [31].
The phaA, phaB and phaC genes may have been acquired through horizontal gene transfer from a recent common ancestor throughout evolution, based on gene order conservation and phylogeny. Considering all these observations, it can be concluded that the PHA biosynthetic genes probably originated through the process of horizontal gene transfer or through native gene relocation.
The majority of the products of surrounding genes in the vicinity of phaA, phaB, and phaC are relatively stable and required for cell survival [32,33]. Our results of gene alignment and phylogenetic analysis show that these genes have been continuously maintained throughout all genomes, meaning that they have been successfully transferred and conserved in the descendants via horizontal gene transfer events for optimal cell machinery function.
After analyzing the results of both gene and protein analysis consisting of sequence alignment and phylogenetic analysis, we hypothesized in this work that during the course of natural selection in evolution in an ancestor, PHA related genes (particularly phaC) were transferred along with surrounding genes and established long-term relationships for maintenance. On the other hand, this can also be a result of horizontal gene transfer in order to improve the genetic stability and maintenance in microorganisms [34].
Conclusion
A study on the gene and protein analysis along with the evolutionary history of PHA biosynthetic genes i.e., phaA, phaB and phaC has shown that horizontal gene transfer and rearrangement of the nucleotide sequences proves to be major contributors for the evolution of PHAs. Gene and protein sequences are not fully conserved across all PHA producing microorganisms but there are conserved nucleotides which indicate that there may have been instances of gene rearrangement due to addition, deletion or replacement of a nucleotide. However, the genes still coded for the relative proteins involved in the biosynthesis of PHAs which shows that even though the sequences are not that much conserved but the functions remain completely conserved. The highest level of conservation was of the protein function than the protein structure followed by the protein and gene sequences. The microorganisms belonging to Proteobacteria appeared to be the closest in terms of the phylogeny studied regarding PHA producers. This indicates that there is more conservation in these microorganisms than those belonging to other families.
References
- Muthusamy MS, Pramasivam S (2019) Bioplastics–an eco-friendly alternative to petrochemical plastics. Curr World Environ 14:49.
- Verma R, Vinoda KS, Papireddy M, Gowda ANS (2016) Toxic pollutants from plastic waste-a review. Procedia Environ Sci 35:701-708.
- North EJ, Halden RU (2013) Plastics and environmental health: the road ahead. Rev Environ health 28:1-8.
- Geyer R, Jambeck JR, Law KL (2017) Production, use, and fate of all plastics ever made. Sci Adv 3:e1700782.
- Madison LL, Huisman GW (1999) Metabolic engineering of poly (3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21-53.
- Anderson AJ, Dawes E (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450-472.
[Crossref] [Googlescholar] [Indexed]
- Ha CS, Cho WJ (2002) Miscibility, properties, and biodegradability of microbial polyester containing blends. Prog Polym Sci 27:759-809.
- Ostle AG, Holt JG (1982) Nile blue A as a fluorescent stain for poly-beta-hydroxybutyrate. Appl Environ Microbiol 44:238-241.
- Murray RGE (1994) Determinative and cytological light microscopy. Methods Gen Mol Biol 7-20.
- Chen GQ (2009) A microbial Polyhydroxyalkanoates (PHA) based bio-and materials industry. Chem Soc Rev 38:2434-2446.
- Troschl C, Meixner K, Drosg B (2017) Cyanobacterial PHA production—Review of recent advances and a summary of three years’ working experience running a pilot plant. Bioengineering 4:26.
[Crossref] [Googlescholar] [Indexed]
- Costa SS, Miranda AL, de Morais MG, Costa JAV, Druzian JI (2019) Microalgae as source of polyhydroxyalkanoates (PHAs)—A review. Int J Biol Macromol 131:536-547.
[Crossref] [Googlescholar] [Indexed]
- Shah AA, Hasan F, Hameed A, Ahmed S (2008) Biological degradation of plastics: a comprehensive review. Biotechnol Adv 26:246-265.
- Zinn M, Witholt B, Egli T (2001) Occurrence, synthesis and medical application of bacterial polyhydroxyalkanoate. Adv Drug Deliv Rev 53:5-21.
- Slater SC, Voige WH, Dennis D (1988) Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-beta-hydroxybutyrate biosynthetic pathway. J Bacteriol 170:4431-4436.
[Crossref] [Googlescholar] [Indexed]
- Schubert P, Steinbüchel A, Schlegel H (1988) Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol, 170:5837-5847.
[Crossref] [Googlescholar][Indexed]
- Chen YJ, Huang YC, Lee CY (2014) Production and characterization of medium-chain-length polyhydroxyalkanoates by Pseudomonas mosselii TO7. J Biosci Bioeng 118:145-152.
[Crossref] [Googlescholar] [Indexed]
- Wang Y, Chung A, Chen GQ (2017) Synthesis of medium‐chain‐length polyhydroxyalkanoate homopolymers, random copolymers, and block copolymers by an engineered strain of Pseudomonas entomophila. Adv Healthc Mater 6:1601017.
- Zeller MA, Hunt R, Jones A, Sharma S (2013) Bioplastics and their thermoplastic blends from Spirulina and Chlorella microalgae. J Appl Polym Sci 130:3263-3275.
- Bhatia SK, Kim JH, Kim MS, Kim J, Hong JW, Hong YG, Yang YH (2018) Production of (3-hydroxybutyrate-co-3-hydroxyhexanoate) copolymer from coffee waste oil using engineered Ralstonia eutropha. Bioprocess and biosystems engineering, 41:229-235.
- Dietrich K, Dumont MJ, Del Rio LF, Orsat V (2017) Producing PHAs in the bioeconomy—Towards a sustainable bioplastic. Sustain Prod Consum 9:58-70.
- Koller M (2017) Advances in Polyhydroxyalkanotes (PHA) production. Bioengineering (Basel) 4:88.
[Crossref] [Googlescholar][Indexed]
- Agustin MB, Ahmmad B, Alonzo SMM, Patriana FM (2014) Bioplastic based on starch and cellulose nanocrystals from rice straw. J Reinf Plast Compos 33:2205-2213.
- Alamgeer M (2019) Polyhydroxyalkanoates (PHA) genes Database. Bioinformation 15: 36-39.
[Crossref] [Googlescholar][Indexed]
- Khan N (2017) MEGA-Core of Phylogenetic Analysis in Molecular Evolutionary Genetics. J Phylogen Evol Biol 5:1-4.
- Price MN, Arkin AP, Alm EJ (2006) The life-cycle of operons. PLoS Genet 2:e96.
[Crossref] [Googlescholar] [Indexed]
- Reams AB, Neidle EL (2004) Selection for gene clustering by tandem duplication. Annu Rev Microbiol 58:119-142.
- Kalia VC, Chauhan A, Bhattacharyya G (2003) Genomic databases yield novel bioplastic producers. Nat Biotechnol 21:845-846.
- Pérez‐Pantoja D, Donoso R, Agulló L, Córdova M, Seeger M, et al. (2012) Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ Microbiol 14:1091-1117.
[Crossref] [Googlescholar] [Indexed]
- Li X, Zhang L, Wang G (2014) Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PloS one 9:e92236.
- Osbourn AE, Field B (2009) Operons. Cell Mol Life Sci 66:3755–3775.
[Crossref] [Googlescholar] [Indexed]
- Gutgsell NS, Deutscher MP, Ofengand J (2005) The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 11:1141–1152.
- Lee WH, Loo CY, Nomura CT, Sudesh K (2008) Biosynthesis of polyhydroxyalkanoate copolymers from mixtures of plant oils and 3-hydroxyvalerate precursors. Bioresour Technol 99:6844-6851.
[Crossref] [Googlescholar] [Indexed]
- Rogozin IB, Makarova KS, Murvai J, Czabarka E, Wolf YI, Tatusov RL, Koonin EV (2002) Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res 30:2212-2223.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi