Research Article, J Appl Bioinforma Comput Biol Vol: 12 Issue: 1
Meta Analysis of Micro RNAs Expressed Under Cadmium Stress In Rice Root Tissue
Kavitha Sankaranarayanan1*, Anitha Mani1and Manonanthini T2
1Department of Biology, Madras Institute of Technology, Anna University, Chromepet, Chennai, India
2Department of Bioinformatics, Madras Institute of Technology, Anna University, Chromepet, Chennai, India
*Corresponding Author: Kavitha Sankaranarayanan
Department of Biology, Madras Institute of Technology, Anna University, Chromepet, Chennai, India
Tel: 9104422232711;
E-mail: skavitham@yahoo.com
Received date: 13 January, 2023, Manuscript No. JABCB-23-68358;
Editor assigned date: 16 January, 2023, PreQC No. JABCB-23-68358 (PQ);
Reviewed date: 30 January, 2023, QC No. JABCB-23-68358;
Revised date: 06 February, 2023, Manuscript No. JABCB-23-68358 (R);
Published date: 13 February, 2023, DOI: 10.4172/2329-9533.1000250.
Citation: Sankaranarayanan K, Mani A, Manonanthini T (2023) Meta Analysis of Micro RNAs Expressed Under Cadmium Stress in Rice Root Tissue. J Appl Bioinforma Comput Biol 12:1.
Abstract
Toxicity due to metal is one the key abiotic stress factor effecting crop yield. Several biological responses take place in plant due to metal stress at levels of both transcription and post transcription. One of the abundant heavy metal pollutants which are highly toxic to both plants and animals is Cadmium (Cd). In plants, miRNAs are associated in regulation of signal transduction, growth and stress responses. Recent studies indicate that miRNAs are involved in both biotic and abiotic stress responses in plants. Meta analysis of miRNAs under Cd stress in different rice root tissue has been done in this study. This study revealed the differential expression of miRNAs under Cadmium stress and their target mRNAs in different root tissues. The study evaluated the expression of miRNAs such as miR812, miR169, miR167, miR166 and miR2118 as the major key players in Cd stress of rice root tissue using statistical measures. The study also involved a detailed literature study and GO enrichment analysis of miRNAs and their target mRNAs using different bioinformatic tools and annotation databases. Also the result of the study indicates that miRNAs and mRNAs could act as a resource for engineering stress tolerance in rice in future studies.
Keywords: Rice, Cadmium stress, miRNA, Meta analysis, Heavy metal toxicity
Introduction
Rice is one of the widely cultivated and consumed crop. Its production is effected by several factors, one among them is heavy metal. Cadmium is one of the most widely found non-essential heavy metal pollutant which has several deleterious effects on both plants and animals at higher concentrations. Cadmium pollution is widely seen in paddy cultivation soils and is easily accumulated in rice, hence Cadmium contaminated rice consumers are prone to Cadmium toxicity. Cadmium pollution is a major threat to Food safety as major source of Cadmium for non-smokers is food and leads to risk of cancer such as in the lung, bladder, breast etc.
MicroRNAs (miRNAs) are defined as 21 nt long small and nonprotein coding RNAs that bind to target mRNAs with complementary sequences in them and regulate gene expression either by mRNA degradation or translational inhibition [1]. Precursor miRNA or pri miRNA has a hairpin like structure that is processed into mature miRNA and is incorporated into RNA Induced Silencing Complex (RISC) which induces the target RNA cleavage [2]. miRNAs in plants are involved in regulating numerous biological processes, like cell identity, developmental patterning, signal transduction and growth [3]. Recently several miRNAs have also been identified to be involved in abiotic and biotic stress responses [4]. The role of miRNAs in plants under abiotic stress was first identified in Arabidopsis [5].
20 miRNAs were first identified in rice in 2004 and their potential targets were predicted. Their target genes were identified to be involved in transcription, disease resistance, transport, metabolism, etc., [6]. 14 novel miRNAs in rice were later found to be associated with various physiological processes [7]. Several miRNAs in rice have been identified, 604 precursors and 738 mature miRNA sequences are there in miRbase version 22.1. Numerous studies are there on miRNA prediction from rice NGS data and conserved miRNAs involved in many abiotic stress responses [8-10].
Heavy metals are extensively used in many industries leading to toxicity and heavy metal pollution. These pollutants have an adverse effect on plant biochemical and physiological aspects and also may result in plant death. It has been identified that in the presence of heavy metals, miRNAs are directly involved in biochemical response by varying the biosynthesis of metabolites containing sulphur that are involved in heavy metal sequestration [11]. Increased sulfur transportation leads to amplified biosynthesis of the precursor for Phytochelatins (PCs) that is Glutathione (GSH) which plays a role in detoxification and sequestration of heavy metal [12]. Thus, under heavy metal stress miRNAs are believed to be involved in the regulation of phytohormone biosynthesis inspite of its direct involvement in the response. Under metal toxicity, miRNAs modify the gene expression in the plant by regulating excess metal complexation process, signal transduction for regulatory biological responses and defense against oxidative stress [13].
In plants like rice, Medicago truncatula and Arabidopsis thaliana a group of Cadmium responsive miRNAs have been recognized [14-16]. 28 novel miRNAs were isolated from rice seedlings treated with Cd. 19 miRNAs were identified in Cd treated rice roots in a microarray based assay. Previously only physiological mechanisms underlying in response to Cd stress were studied. In a study of gene regulation in rice under Cd stress, it was found that genes associated with heat shock proteins, cytochrome P450 family proteins, glutathione Stransferase, protein kinases, transcription factors and few transporter were induced [17].
Under various stress conditions, transcriptome profiling of miRNAs has been reported widely. Many studies have also stated that miRNA target expression and/or degrade data to identify the role of miRNAs associated with heavy metal stress [18,19]. With advances in genomics and high throughput sequencing technologies, the number of miRNAs identified in plant genome continues to increase. There are still several novel miRNAs to be identified which are associated with either stress response or regulation of the developmental process.
This study has been done to acquire a deep understanding of the miRNA expression profile under Cadmium stress in different tissues (Vascular Bundle (VB), Exodermises Plus epidermis (EP) and Cortex (Cor)) of rice root. In the present study meta analysis of the expression of miRNAs observed under Cadmium stress in rice root tissues was performed by using publicly available microarray gene expression data.
Materials and Methods
Data mining
The present study is a bioinformatics approach to identify the miRNAs that are widely expressed under Cadmium stress in rice root. NCBI GEO has a huge amount of microarray data deposited by several authors, which can be easily downloaded and processed to find new insights using meta analysis [20]. GEO of NCBI was used to load microarray gene expression profile data using GEO accession number GSE40549 [21]. There are a total of 39 samples out of which, 3 samples each were selected from Vascular Bundle (VB), Exodermis Plus epidermis (EP) and Cortex (Cor) under Cadmium stress condition (10 μM CdCl2 added for 24 hrs after 33 days after germination) and 3 each of the above under control condition.
Data analysis
The microarray dataset from GEO was downloaded and was analyzed using GEO2R tool. GEO2R is a simple web based tool that helps to compare two or more groups of samples in a GEO datasets to analyses and identify differentially expressed genes among different experimental conditions. GEO2R is a bioconductor based tool used to compare GEO datasets submitted originally which were then processed with the help of GEO query and limma bio conductor package. Bio conductor is an open source package based on the R programming language that offers software tools and programs for the analysis of genomic data. GEO2R utilizes the ANOVA statistical testing on grouped GEO datasets. The resulting tables with list of genes are ordered by expression values (Fold change) and are sorted based on p-values. There are options to select different columns and save the results. There are several options to choose different algorithms and to set the parameters. In this study, Benjamini and Hochberg algorithm was used for analysis. The result tables with large number of genes were carefully analyzed as majority of them are irrelevant. After analysis, based on p-value<0.001, genes are selected for further analysis. Log2 fold change (logFC) values were obtained from GEOR and were used to identify the differentially expressed miRNAs in the respective tissue. Using BLASTN search against the miRNA database, miRBase 22.1 conserved miRNAs were identified [22,23].
miRNA target prediction and functional annotation
The analyzed miRNAs were then used for predicting mRNA targets. For predicting probable targets of miRNAs, the miRNA target interactions were identified using the miRNA id’s retrieved from miRBase using degradome based plant MiRNA target interaction and network database DPMIND. For the verified miRNA targets, the network was generated using DPMIND.
Here, potential targets of miRNAs in all three tissues were subjected to go functional enrichment analysis using the Panther database. PANTHER is a part of the gene ontology reference genome project intended to categorize proteins and their genes for high throughput genomics data analysis. The results for lists of miRNAs and their target genes related to a specific protein family/subfamily, GO molecular function, GO biological process, GO cellular processes and pathway were generated in the form of table and bar graphs to fully understand the function of these targets.
Results
The results were further analyzed for relevant miRNAs using p value<=0.001 as the cut off value and 274 miRNAs were identified in total to be differentially expressed under Cd stress in root tissue. Table 1 shows the top 10 miRNAs expressed under Cd stress in various tissues in rice root. Out of 274 miRNAs 180 were specific to EP, 19 to VB and 23 to Cor. Out of 274 miRNAs only 4 were common between all the three tissue types, whereas 21 miRNAs were common between EP-VB, 3 between VB-Cor and 24 between Cor-EP and is depicted in the venn diagram (Figure 1). Based on Venn diagram analysis, we analyzed differences and crosstalk of miRNA expression among VB, Cor and EP in rice root.
Table 1: Up and down regulated miRNA, their logFC and P value in rice tissue under Cd stress.
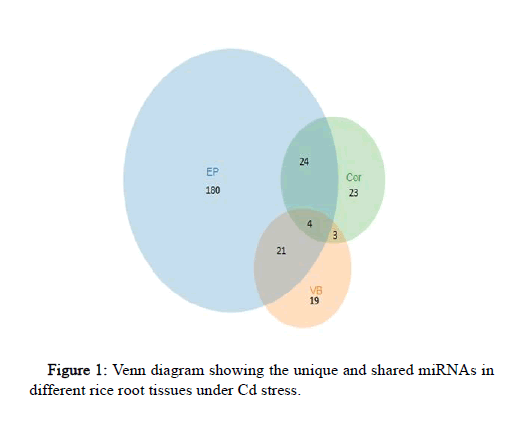
Figure 1: Venn diagram showing the unique and shared miRNAs in different rice root tissues under Cd stress.
miRNA target prediction
It is very important to identify miRNA targets for understanding miRNA function. Using dpmind probable miRNA targets were identified in order to understand further about the role of the miRNAs under Cadmium stress. In Cor, 813 targets were identified, out of which 253 were specific to the cortex. Whereas in EP, 2508 targets were identified, out of it 1205 were specific to EP. In VB, 480 targets were identified with 109 specific to VB. Overall 99 targets were identified to be common in all the three different tissues.
Functional annotation of miRNA targets
Under metal stress, several signaling pathways are activated which are involved in protecting the plant. To understand the role of target genes that are Cd treatment responsive in the rice root tissue, their Gene Ontology (GO) analysis was performed.
Here, probable targets of miRNAs were studied using panther database for go functional enrichment analysis. Panther database predicts all go like biological process, molecular function, and cellular component terms and it also provides pathway information that can be exported either as pie or bar chart. Graphical results were generated to completely understand the role of these targets. Using the panther database, identified targets were grouped based on their molecular function, biological processes and cellular compartment. This grouping helped to determine the distribution of the miRNA targets, according to molecular function (Figure 2), biological process and cellular compartment (Figures 3 and 4). Cor cluster with the biological process comprised proteins that are involved in biological regulation (GO:0065007), localization GO:0051179), metabolic process (GO: 0008152) and response to stimuli (GO:0050896). This analysis clustered the targets into three molecular functional classes which were related with binding (GO:0005488), catalytic activity (GO:0003824) and transcriptional regulator activity (GO: 0140110). The target groups were then further divided into three subcellular distributions according to the cellular compartment. Vb cluster with the biological process comprised proteins that are involved in cellular process (GO:0009987), metabolic process (GO: 0008152) and multicellular organismal process (GO:0032501). This analysis grouped the targets into a single molecular functional class which is related to catalytic activity (GO:0003824). The targets were further categorized into single subcellular distribution according to the cellular compartment. While EP cluster with the biological process comprised proteins that are involved in biological regulation (GO:0065007), cellular component organization or biogenesis (GO:0071840), cellular process (GO: 0009987), localization (GO:0051179) and metabolic process (GO: 0008152). This analysis clustered the targets into six molecular functional classeswhich were associated with binding (GO:0005488), catalytic activity (GO:0003824), structural molecule activity (GO:0005198), transcriptional regulator activity (GO:0140110), translation regulator activity (GO:0045182) and transporter activity (GO:0005215). The targets were further categorized into five subcellular distributions according to the cellular compartment.
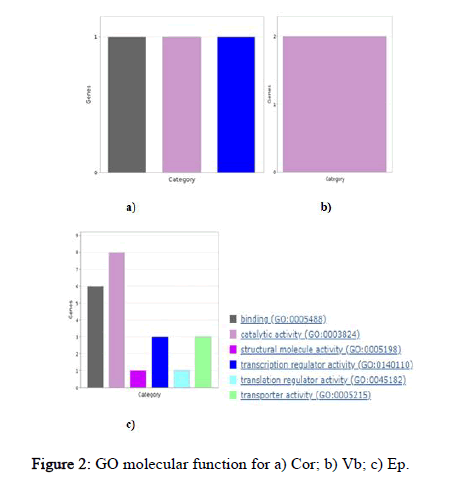
Figure 2: GO molecular function for a) Cor; b) Vb; c) Ep.
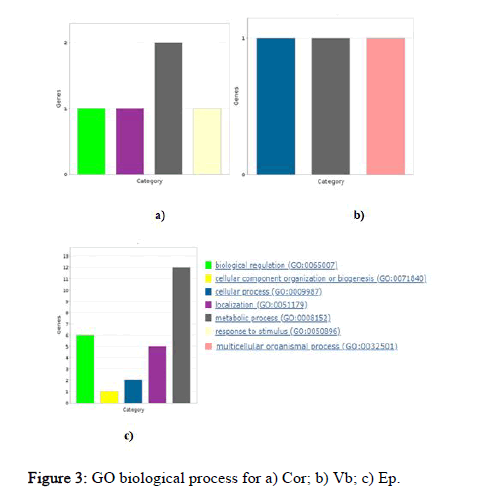
Figure 3: GO biological process for a) Cor; b) Vb; c) Ep. Figure
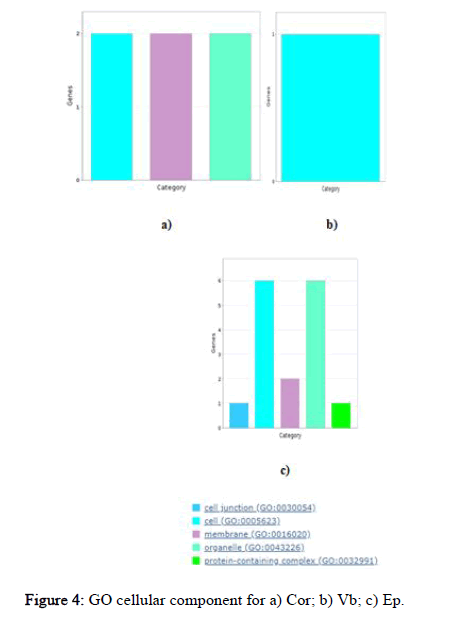
Figure 4: GO cellular component for a) Cor; b) Vb; c) Ep.
Discussion
Stress due to heavy metal is one of the major abiotic factors influencing plant growth. Cd is a non-essential element which even at low concentrations is toxic. Plant response to Cd stress is complex and involves several physiological, morphological and molecular changes that may lead to adaptive changes or have a deleterious effect on the plant. From previous studies, it can be understood that miRNAs are considered as one of the vital gene regulators. With rising number of studies about the role of miRNAs in crop plants, a lot of progress has been made in analyzing and characterizing the plant miRNAs. miRNAs play a vital role in several plant processes like hormone signaling and signal transduction pathways, growth and development during both vegetative and reproductive stages and in maintaining homeostasis under abiotic and biotic stress [24].
In the present study, meta analysis of miRNA’s expressed in three different tissues of rice root under Cd was done. For that gene expression data were collected from the 3 publicly available data set. Totally 274 miRNAs were found in rice root tissue under Cd stress using miRBase which belonged to 50 miRNA families. In EP, 229 miRNAs were identified out of which 180 were specific to EP only. Using logFC ≥ 1.5 as the cut off, it was identified that out of the total 229 miRNAs in EP, 201 (87.8%) were up regulated. Similarly, in Cor out of the total 54 miRNAs, 39 (72.2%) were up regulated and 15 (27.8%) were down regulated. In VB, out of the total 47 miRNAs, 23 (48.9%) were up regulated and 24 (51.1%) were down regulated. Among the detected miRNA’s in Cor, the miR812 followed by miR169 1 families comprised the highest number with 5 and 4 members. In VB, among the detected miRNA’s the miR812 and miR166 families comprised the highest number with 3 members each. Similarly, in EP the miR812 followed by miR169, miR2118 and miR167 families comprised the highest number with 16, 9, 9, and 6 members respectively. Previous studies in rice plant have reported that miR812, miR169, miR167, miR166 are regulated under metal stress. miR812 was predicted to target glycosyl hydrolase family 17, mir169 was found to target Nuclear transcription Factor Y subunit (NFY), miR167 to target Auxin Response Factor (ARF) and miR166 to target START domain containing protein. By regulating ARF, miR167 thereby regulates auxin signaling and thus several developmental processes in rice plant under metal stress. In rice, auxin induces the expression of GH3 genes which play a key role in defense responses. miR2118 was predicted to target protein kinase associated with protein phosphatase thus regulate signal transduction in rice plant under metal stress of the predicted miRNAs, 16 miRNAs were found to have no potential verified or predicted target when studied using DPMIND. Table 2 indicates the list of non-annotated miRNAs in this study. Osa MIR 5153, 2919 and 11340 were found to be down regulated in VB, while osa-MIR5077 was down regulated in Cor. Osa MIR5153, 2932, 11340, 417, 1430, 2925, 5805, 438, 2879, 5493, 11341 and 7692 were found to be up regulated in EP. These miRNAs can be further studied in order to understand their role in Cd stress in rice root.
Table 2: List of non-annotated miRNAs.
Gene ontology analysis for miRNA targets was carried out for a better understanding of the functions of miRNAs. GO enrichment analysis helps in identifying the role of target genes as in molecular function, biological process and cellular component involved in the response to various stress condition. On the basis of GO enrichment analysis, these entire miRNA target genes were mainly distributed in biological processes, ‘molecular function’ and ‘cellular component’ under Cd stress. This suggests that wide ranges of complex responsive approaches are initiated to adapt to external Cd stress in rice. The functional annotation of the target genes common in Cor tissue showed that most of the genes encode for binding, catalytic activity and transcriptional regulator activity proportionally. Whereas in EP it showed that most of the target genes encode for catalytic activity followed by binding and equally for transcription regulator activity and transporter activity and minimally in structural molecule activity and translation regulator activity. While the functional annotation of the target genes in Vb showed only catalytic activity. The previous study of GO analysis of Cd stress regulated genes in root indicated gene enrichment in stress response and metabolic processes, while in shoot only genes involved in lipid metabolism, indicating the major difference in the processes that take place in two different tissues under Cd stress.
Conclusion
With increasing studies indicating the importance of miRNAs in crop plants, thus there is an increased need for further studies for improving the agronomic traits in crop plants. The present study revealed the differentially expressed miRNAs in different root tissues under Cadmium stress, which may be useful for studying their differential expression during root growth and under other stress conditions more precisely. GO enrichment aided in revealing the involvement of miRNAs and their targets in diverse developmental processes. In conclusion, this study revealed a statistical framework of miRNAs in rice root tissue under Cd stress. Also, this study of differentially expressed genes could act as a resource for potential genes for engineering stress tolerance in rice.
Acknowledgements
AM would like to thank Anna University for providing Anna Centenary Research Fellowship during the research work.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116:281-297.
- Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19-53.
- Tang M (2014) Integrated analysis of miRNA and mRNA expression profiles in response to Cd exposure in rice seedlings. BMC Genomics 15:835.
- Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14:836-843.
- Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell, vol. 16, pp. 2001-2019.
- Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH (2004) Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res 32:1688-1695.
[Crossref] [Googlescholar][Indexed]
- Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNA from rice. Plant Cell 17:1397-1411.
[Crossref] [Googlescholar][Indexed]
- Sunkar R, Chinnusamy V, Zhu JH, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12:301-309.
- Zhang Q (2007) Strategies for developing green super rice. Proc Natl Acad Sci U S A 104:16402-16409.
- Mittal D, Sharma N, Sharma V, Sopory SK, Sanan-Mishra N (2016) Role of microRNAs in rice plant under salt stress. Ann App Biol 168:2-18.
- Na G, Salt DE (2011) The role of sulfur assimilation and sulfur-containing compounds in trace element homeostasis in plants. Environ Exp Bot 72:18-25.
- Yang ZM, Chen J (2013) A potential role of microRNAs in plant response to metal toxicity. Metallomics 5:1184-1190.
[Crossref] [Googlescholar] [Indexed]
- Gielen H,Remans T, Vangronsveld J, Cuypers A (2012) MicroRNAs in Metal Stress: Specific Roles or Secondary Responses?. Int J Mol Sci 13:15826-15847.
- Huang SQ, Peng J, Qiu CX, Yang ZM (2009) Heavy metal-regulated new microRNAs from rice. J Inorg Biochem 103:282-287.
[Crossref] [Googlescholar][Indexed]
- Ding Y, Chen Z, Zhu C (2011) Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot 62:3563-3573.
- Zhou ZS, Huang SQ, Yang ZM (2008) Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochem Biophys Res Commun 374:538-542.
[Crossref] [Googlescholar][Indexed]
- Ogawa I, Nakanishi H, Mori S, Nishizawa NK (2009) Time course analysis of gene regulation under cadmium stress in rice. Plant Soil 325:97-108.
- Zhou ZS, Song JB, Yang ZM (2012) Genome-wide identification of Brassica napusmicro RNAs and their targets in response to cadmium. J Exp Bot 63:4597-4613.
- Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2012) Genome-wide identification of Medicago truncatula micro RNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35:86-99.
- Barozai MYK, Wahid HA, (2012) In silico identification and characterization of Cumulative abiotic stress responding genes in Potato (Solanum tuberosum L.). Pak J Bot 44:57-69.
- Ogo Y, Kakei Y, Itai RN, Kobayashi T, Nakanishi H, et al. Spatial transcriptomes of iron-deficient and cadmium-stressed rice. New Phytol 201:781-94.
- Kozomara A, Griffiths-Jones S (2011) miRBase: integrating micro RNA annotation and deep-sequencing data. Nucleic Acids Res 39:D152-D157.
- Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids 42:D68-D73.
- Sun G, Stewart CN, Xiao P, Zhang B (2012) MicroRNA expression analysis in the cellulosic biofuel crop Switchgrass (Panicum virgatum) under abiotic stress. PLoS ONE 7:e32017.
[Crossref] [Googlescholar] [Indexed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi 
