Research Article, J Appl Bioinforma Comput Biol Vol: 12 Issue: 1
Molecular and Phylogenetic Analysis of Inducible Nitric Oxide Synthase among Different Organisms Molecular and Phylogenetic Analysis of Inducible Nitric Oxide Synthase among Different Organisms
Arti Sharma1*, Jyoti Parkash2, Priya Godara2, Nymphaea Arora2, Vikash Prashar2, Tania Aror2, Randeep Singh2, Arpita Banerjee2 and Harish Changotra3
1Department of Computational Biology, School of Basic Sciences, Central University Punjab, Ghudda, Bathinda 151 401, Punjab, Bathinda, India
2Department of Zoology, School of Basic Sciences, Central University Punjab, Ghudda, Bathinda 151 401, Punjab, Bathinda, India
3Department of Molecular Biology and Biochemistry, School of Life Sciences, Guru Nanak Dev University, Amritsar 143005, Punjab, India
*Corresponding Author: Arti Sharma
Department of Computational Biology, School of Basic Sciences, Central University Punjab, Ghudda, Bathinda 151 401, Punjab, Bathinda, India
Tel: 7986980164;
E-mail: jyoti.parkash@cup.edu.in
Received date: 18 January, 2023, Manuscript No. JABCB-23-61356;
Editor assigned date: 20 January, 2023, PreQC No. JABCB-23-61356 (PQ);
Reviewed date: 03 February, 2023, QC No. JABCB-23-61356;
Revised date: 10 February, 2023, Manuscript No. JABCB-23-61356 (R);
Published date: 17 February, 2023, DOI: 10.4172/2329-9533.1000246.
Citation: Sharma A, Parkash J, Godara P, Arora N, Prashar V, et al. (2023) Molecular and Phylogenetic Analysis of Inducible Nitric Oxide Synthase among Different Organisms Molecular and Phylogenetic Analysis of Inducible Nitric Oxide Synthase among Different Organisms. J Appl Bioinforma Comput Biol 12:1.
Abstract
Nitric Oxide (NO) is a hetero-nuclear diatomic molecule which is formed from L-arginine through catalytic activity of Nitric Oxide Synthase (NOS). NO is an important messenger molecule known to be involved in dilation of smooth muscles, penile erection, synaptic plasticity. The three isoforms of NOS are nNOS (neuronal NOS), iNOS (inducible NOS), and eNOS (endothelial NOS). The iNOS is coded by NOS2 gene and is known to play an important role in body’s immune response involving inflammatory response, leukocyte mediated microbial destruction. In this study the iNOS sequences was compared among 18 organisms from arthropods to mammals including bacteria. To understand the implication of iNOS the domain as well as phylogenetic analysis was done. The various biochemical, physico-chemical, and nearly two possible transmembrane models were predicted to know more about the functional regions and structural features of iNOS. One deleterious SNP was predicted through literature survey which is known to have harmful impact on the function of protein and may be used for genomic association studies. The 14 different Post-translational Modifications (PTMs) were predicted which might play an important role in influencing protein interaction network. Nearly 8 Transcription Factors (TFs) were predicted in mouse and rat which may directly influence the NOS2 expression. Using different in silico methods we actually made a comparative analysis of the different parameters of iNOS to deeply understand more of its regulatory activities. In conclusion, results obtained from this study may be used to create a link between the regulations of iNOS in various organisms and in future the results might be exploited for the clinical trials to regulate various autoimmune disorders.
Keywords: Nitric Oxide Synthase (NOS) 2; Single nucleotide po
lymorphisms; Post translational modifications; Transcription
factors; Nitric oxide; Autoimmune disorders
Introduction
Nitric oxide synthases are a family of enzymes that catalyze the Nitric Oxide (NO) production from L-arginine in the presence of O2, Nicotinamide Adenine Dinucleotide Phosphate (NADPH), Flavin Adenine Dinucleotide (FAD), Flavin Mononucleotide (FMN) [1]. The three isoforms of NOS are coded by separate genes as neuronal NOS coded by NOS1, inducible NOS by NOS2, and endothelial NOS by NOS3. Early roles of NO in the dilation of the smooth muscles were known but recent discoveries have proved its important roles in inflammatory responses, leukocyte mediated microbe destruction, penile erection and release of Luteinizing Hormone Releasing Hormone (LHRH) [2]. Nitric oxide produced by inducible NOS (iNOS) in the immune system is a cytotoxic agent [3]. Inducible NO synthase (iNOS) is expressed after cell activation and NO is produced for long periods of time (hours to days) compared to other isoforms [4]. The iNOS has been described as calcium insensitive, likely due to its non-covalent interaction with Calmodulin (CaM). The iNOS activity is mostly regulated at transcriptional level and through its intracellular distribution. NO immediately diffuses across the biological membranes following its synthesis as it is not stored in the vesicles. However to exert an action is delineated in the NO by its half-life and also by the proximity of NO containing cells to their target which must be between 200 μm [5,6]. It has been reported that the receptor of NO is the Soluble Guanylate Cyclase (sGC) which results in the formation of cGMP to mediate intracellular effects when activated by nitric oxide [7].
NOS2 gene found on human chromosome 17 q 11.2-12 with 37 kb length comprises 1153 amino acid sequence and has molecular weight of 131 kDa. Human iNOS gene comprises 27 exons with transcription start site in exon 2 and stop codon in exon 27. Exons 1-13 encode oxygenase domain and 14-27 codes for reductase domain. C-terminal reductase domain of NOS2 binds to NADPH which transfers electrons to FAD, and then to FMN and finally to N-terminal domain. This Nterminal oxygenase domain contains a heme, arginine binding sites, tetrahydrobiopterin and calmodulin.
A very high level of nitric oxide is secreted by the induced macrophages which cause the breaking and fragmentation of DNA strands in the target cells. Due to the more affinity towards the iron which is protein bound, NO may inhibit enzymes which are known to contain iron into their catalytic centers which may include iron sulphur cluster depending enzymes (complexes I and II) known to be involved into the mitochondrion electron transport. Interestingly, the nonimmune cells can also be induced by the cytokines to secrete the NO causing tumor cell lysis. It is reported that the tumor and immune system interact with each other through macrophages and for the activation of M1 macrophages iNOS is the surrogate marker [8]. Excessive high levels of nitric oxide secreted by the iNOS may result in septic shock and atherosclerosis [9,10]. Studies have shown that ghrelin hormone alters the activity of iNOS. A recent data from the rat heart suggests the alteration of NOS2 activity due to ghrelin hormone as ghrelin might activate IGF-1 (insulin like growth factor-1) pathway involving interaction with Akt/ERK 1/2 signaling pathways [11,12]. Evidences have shown that members of MAPK family are important modulators of NOS2 gene expression in the multiple cell types [13].
Compared with the wild types, the maximum activity of NO is retained by the deletion mutants at free calcium ions in lower concentration [14]. According to some case control and genomic association studies there exists a strong linkage between single nucleotide polymorphisms (SNPs) in NOS2 gene and the occurrence of rheumatoid arthritis, osteoarthritis, muscular dystrophy, gastric cancer, cardiovascular diseases, diabetes, tuberculosis, malaria and bacterial meningitis [15]. It has been observed that during the conditional knock out of NOS2 gene in mouse model (CVN-AD), a great alteration in the NO mediated effects were seen which were related to the AB-production and deposition of amyloid leading to Alzheimer’s disease and producing more human like responses [16]. In another study the knockout of NOS2 gene in mice resulted in the increased Barbering Behavior (BB) that is generally seen in the laboratory animals and is a model for Trichotillomania (TTM) which is an impulse control disorder. It suggests that the maturation of the cortical neurons would be affected by the lack of NOS2 expression in the early stages which may result in the described phenotype [17]. The post mortem reports have claimed the presence of iNOS protein in the brains of patients with neurodegenerative diseases including Parkinson’s and Alzheimer’s disease [18]. The growth of the tumor cells suppresses the host’s immunity which is supported by the fact that during their proliferation, the tumor cells secrete the nitric oxide which is known to inhibit the proliferation and finally apoptosis of the T-lymphocytes [19]. An effective DNA repairing system and the expression of the heat shock proteins (HSPB1 and HSPA1A) make the human cells more resistant towards the harmful effects of NO [20]. It has been reported that the co expression of COX-2 (Cyclooxygenase-2) and NOS2 enhances the growth of tumor and in turn reducing the survivability in the estrogen receptor negative breast cancers [21]. COX-2 and NOS2 are indicators with strong prognostics in the patients with triple negative breast cancer. The presence of iron-heme group in COX-2 suggests its direct interaction with the NOS2 [22]. It has been reported that COX-2 induces angiogenesis which is essential for tumor growth and its activity is enhanced by Nitric Oxide (NO) produced by NOS2. NO and COX-2 produce mediators that regulate cellular growth in tumors which may be mediated by formation of peroxinitrate and prostanoids involving cross-talk between the two enzyme systems. Recent data suggested that translational blockage is produced in gene expression of hiNOS (human NOS) by miRNA (miR-939) which binds to 3’UTR. The increase in miR-939 and iNOS transcription is induced by cytokines which may lead to inhibition in translation in a type of check and balance system suggesting a dual regulation of NOS2 expression [23]. It was demonstrated that NLR family CARD domaincontaining protein 4 (NLRC4)/Caspase-1 plays a very important role in regulating chromatin accessibility of NOS2 promoter by involving poly ADP-ribose polymerase-1(PARP 1) cleavage [24]. The binding of STAT5B in the promoter element (gamma-interferon-activated element) of the NOS2 gene leads to the transcription of NOS2 mRNA resulting into the protein expression in the pancreatic β-cell lines in mouse. The mutated STAT5B cause the silencing of NOS2 gene which leads to the accumulation of H2O2. The resultant oxidative damage causes the β-cell damage [25]. The low level expression of NOS2 cause the over expression of mutant p53 protein which may lead to over expression, cell proliferation in excess and finally result in the progression of the hepatocellular carcinoma. EIK-3, member of ETS (E26 transformation specific) transcription factor family has been proved as a strong repressor of NOS2 promoter and hence may serve in regulation of NOS2 gene. The uncoupling of NOS2 and decreased levels of tetrahydrobiopterin (BH4) cause arterial oxidative stress and lead to chronic heart failure.
By considering the functional relevance and the significance in the clinical aspects of NOS2, a necessity generates for computational characterization of it to deeply understand more about the characteristics which are less studied that might otherwise be time consuming and a tedious work. Additionally, the in-silico analysis of NOS2 may be of great help for its implications in the population studies. The data provided by this study would help to unravel various functions of NOS2. In this paper, we actually tend to compile the data based on different parameters from different species starting from arthropods to mammals including prokaryotes (bacteria) to study the interactions of iNOS with other molecules/factors and provide a link between them. In future this may be exploited for the clinical trials in the treatment of various disorders especially the autoimmune diseases.
Materials and Methods
The computational methodology which is followed in the study is represented in the form of a flow diagram (Figure 1) which includes the 9 main steps that are as follows:
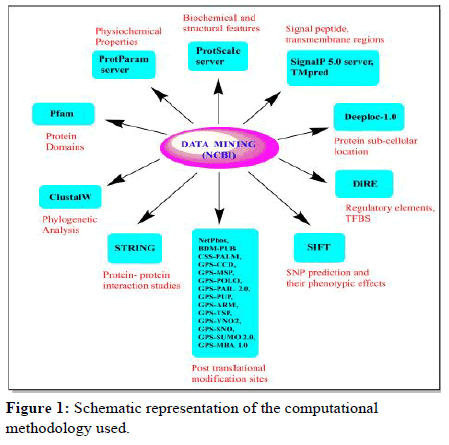
Figure 1: Schematic representation of the computational methodology used.
• Domains analysis,
• Phylogenetic analysis,
• Sequence feature based analysis,
• Detection of signal peptides and transmembrane regions,
• Protein sub cellular localization,
• Identification of the regulatory elements and overrepresented TFBS,
• Prediction of Single Nucleotide Polymorphism (SNP) with their phenotypic effects,
• Identification of post-translational modification sites,
• Protein-protein interaction studies.
Data mining
The sequence of protein for human NOS2 was retrieved from the National Center for Biotechnology Information with uniprot KB ID: P35228 and gene ID: 4843. The protein sequence was taken in FASTA format. The human protein sequence was used as query sequence to deeply understand the NOS2 gene. The protein sequences of 17 other species from arthropods to mammals including bacterial sequence were retrieved from Gene Bank which compares sequence of our interest with the available sequences in database considering maximum similarity and lower e-value. The bacterial oxygenase and reductase sequences were taken separately as no complete bacterial sequence was found.
Phylogenetic analysis
Pfam database provides an accurate and complete classification of the protein families along with their domains. Identifying the domains present in the protein helps us to accurately and deeply understand the functions of that protein. Protein sequences of NOS2 gene across all the organisms under study were investigated for domain analysis in Pfam. For the analysis of evolutionary relationship, NOS2 protein sequence for the human and other species obtained were provided as an input and then multiple sequence alignment and the phylogenetic tree construction was done using Clustal Ω. Clustal Ω uses seeded guide trees and HMM profile-profile techniques to generate multiple sequence alignments. For the phylogenetic reconstruction the Neighbor Joining (NJ) method with 1000 bootstrap value was applied.
Sequence feature-based analysis
It includes various biochemical, physiochemical features including the structural properties that might help to unravel its various characteristics which in turn help in the analysis of protein functions, characters and the protein-protein interactions. For this ProtParam and Prostate were used and protein sequences were provided as an input. ProtParam was used for computing various physiochemical properties which include instability index, molecular weight and Grand Average of Hydropathicity (GRAVY), atomic composition, aliphatic index, amino acid composition, estimated half-life and theoretical pI. ProtScale was used to compute biochemical and structural features which include hydrophobicity, bulkiness, accessibility, mutability, polarity and flexibility that play an important role to retain protein structure and its stability.
Identification of signal peptides and the transmembrane regions
Signal P-5.0Server was used for prediction of Signal peptide (SP), location of their Cleavage Sites (CS) and others (when sequences do not have any kind of signal peptide). The Signal P-5.0 server can distinguish between three different types of signal peptides in Bacteria and Archaea. Including the conditional random field, Signal P-5.0 is also based on the deep convolutional and architecture of recurrent neural network. The TMpred program was used to make predictions of membrane spanning regions along with their orientations. The algorithm is based on the statistical analysis of TMbase which is a database of naturally occurring transmembrane proteins. The prediction is made by using a combination of several weight- matrices for scoring. DeepLoc-1.0 was used to predict the location of iNOS, as it can differentiate between 10 different localizations: Cytoplasm, Endoplasmic reticulum, Peroxisome, Lysosome/Vacuole, Extracellular, Golgi apparatus, Cell Membrane, Chloroplast, Mitochondrion and Nucleus.
Identification of distant regulatory elements and transcription factor with their binding sites (TFBS)
DIRE (Distant Regulatory Elements of co-regulated genes) was used for the identification of the Regulatory Elements (REs) and Transcriptional Factor with their Binding Sites (TFBS). Identification of REs and TFs enhance functional annotation of genome that lead to better understanding of gene regulation. Regulatory elements in NOS2 gene were identified with top 3 ECRs+promoter ECRs as target elements and random set of 5000 genes as background/control genes (default settings).
Prediction of non-synonymous single nucleotide polymorphisms
SIFT (Sorting Intolerant from Tolerant) was used for predicting whether an amino acid substitution affects the protein function based on amino acid physical properties and sequence homology. This database can be applied to both naturally occurring non-synonymous polymorphisms as well as laboratory-induced missense mutations. It gives SIFT score, SIFT median, number of sequences at position, SIFT prediction and many other relevant information related to the Single Nucleotide Polymorphisms (SNPs). SIFT score value ranges from 0 to 1 and amino acid substitution is predicted if score is ≤ 0.05 and considered tolerated if score is Ë?0.05. Median score value has range between 0 to 4.32, ideally between 2.75 and 3.5. This can be used for measuring diversity of sequences for prediction. A warning will occur if it is Ë?3.25 indicating that prediction was based on closely related sequences. Sequence at position shows number of sequences that have an amino acid at the position of prediction. If the substitution is located at beginning or end of any protein, there may be only a few sequences represented at that position. SIFT prediction shows whether the SNP is tolerated or deleterious.
Elucidation of post-translational modification sites
Post translational modifications are studied in order to characterize the regulation of gene expression. It defines the fate of proteins after the process of translation in eukaryotic cells. Hence the prediction of PTM sites is useful in study of diagnosis of various diseases. PTM sites includes BDM-PUB for ubiquitination, GPS-MSP for methylation, CSS-Palm for palmitoylation, GPS-SUMO for sumoylation, GPS-SNO for S-nitrosylation, GPS-YNO2 for tyrosine nitration, GPS-CCD for calpain cleavages, GPD-Polo for Polo like kinases, GPS-PUP for pupylation, GPS-MBA for MHC-binding, GPSARM for APC/C recognition motif, GPS-TSP for tyrosine sulfation, GPS-PAIL for lysine acetylation and NetPhos for phosphorylation.
Protein-protein interaction studies
STRING version 11.0 is used for generating protein–protein interaction network that helps in the system level of understanding of the cellular processes. Such networks might be very useful for analyzing functional, structural and evolutionary properties of the proteins. The exploration of protein-protein interactions may put forward new directions in the future experimental research and prediction of cross species for an efficient interaction mapping.
Results
Data retrieval
The amino acid sequence of human NOS2 was obtained from NCBI and used as query sequence against Gene Bank. The sequences of 17 other species from arthropods to mammals including bacteria were retrieved depending upon the lowest e-value. These sequences were identified as putative NOS2 gene which was further confirmed by conserved domain prediction. These NOS2 protein sequences from 18 different families were analyzed and compared using various in silico tools. The list of species under study along with their IDs and common names used are listed in supplementary Table 1.
Table 1: The comparative values of the ProtParam parameters in the organisms under study.
The Pfam database provides us accurate and complete classification of the protein families and domains. The presence of different domains in varying combinations in different proteins give rise to diverse repertoire of proteins found in nature. Pfam is a collection of protein domain families with each family represented by multiple sequence alignments and hidden Markov model. The results show that there are four domains in human. The oxygenase domain lies at aligned region 137-497 while it has no clan. Flavodoxin domain lies at aligned region 541-672 and it has clan CL0076 which has 5 families i.e. FAD binding 1, FAD binding 6, FAD binding 8, FAD binding 9, LUM binding. FAD binding domain lies at aligned region 725-947 and it has clan CL0042 which comprises of 7 member families i.e. flavodoxin 1, flavodoxin 2, flavodoxin 3, flavodoxin 4, flavodoxin 5, flavodoxinNdr l, FMN red. The oxidoreductase NAD binding domain lies at aligned region 979-1093 and it has clan CL0091 which has 4 families i.e. Colicin M, NAD binding 1, NAD binding 6, SIP. As complete sequence of bacteria was not available so bacteria oxygenase and reductase sequences were taken separately. Oxygenase sequence showed only the oxygenase domain while reductase sequence showed flavodoxin, FAD binding and oxidoreductase NAD-binding domains. Rabbit shows only FAD binding domain at aligned region 96-318, and oxidoreductase NAD-binding domain at aligned region 350-464. The domain positions of all the species are shown in Table 2.
Table 2: The comparative values of ProtScale parameters in the organisms under study.
Phylogenetic analysis exhibits evolutionary links of NOS2 disclosing its conservation profile
Multiple sequence alignment of organisms under study was done using Clustal Ω using its default settings which makes use of seeded guide trees and the HMM profile-profile techniques. It helps us in inferring the presence of ancestral relationships between the sequences of different organisms. It was seen that sequence was conserved from aplysia to dog while bacteria (oxygenase) shows no similar conserved sequences. The phylogenetic tree using neighbor- joining (N-J) method obtained represents evolutionary links among 18 different species from arthropods to mammals, also including prokaryotic (bacteria) sequence (Figure 2).
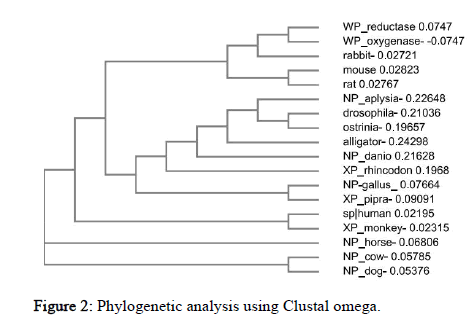
Figure 2: Phylogenetic analysis using Clustal omega.
From the tree it could be observed that drosophila and ostrinia, human and monkey, gallus and pipra, cow and dog, mouse and rat are more closely related to each other. Surprisingly, Rabbit and bacteria belong to the same clad. Horse is an out-group which is the most distant related species in the cladogram in terms of NOS2 sequence and functions as a point of comparison and reference group. Similarly, the relatedness of all other species could be seen from the tree.
NOS2 is found to be a hydrophilic protein
The ProtParam result includes GRAVY (Grand average of hydropathicity) which is of negative value in all the species. Hence, NOS2 is hydrophilic in all species. Protein is considered stable if instability index value is less than 40. The Aliphatic index which is defined as the relative volume occupied by aliphatic side chains is considered one of the positive factor for the increment in thermo stability of globular proteins. The in vivo half-life is defined as the prediction time taken by half amount of protein to disappear after its synthesis in a cell. The Extinction coefficient indicates the amount of light absorbed by protein at certain wavelength. Two values have been produced by the ProtParam based on Extinction coefficients for proteins measured at 280nm with the first one showing the computed value based on assumption that all cysteine residues appear as cystines and the second one assuming that no cysteine appears as half cystine. The comparative values of the ProtParam parameters of all 18 species under study are listed in Table 1. From the table we observe that value of instability index (II) is greater than 40 in all the organisms except bacteria reductase which has II value of 33.28, describing protein as stable. Similarly, the Aliphatic index value is highest in bacteria reductase (92.13) and lowest in aplysia (74.68), which describes reductase as most thermostable. The ProtScale was used for analyzing hydrobhobicity, bulkiness, accessibility, mutability, polarity and flexibility. The hydrophobicity directs protein-protein interactions influencing folding of protein and side chain packing of amino acid and so determines the protein stability. The dipole-dipole interactions between positively and negatively charged residues represent polarity. The probability of the protein to change during evolution is shown by relative mutability. The free transfer energy from inside to the outside of globular proteins is represented by accessibility. Flexibility helps in the building of the interactions of protein with other proteins, ligands or nucleic acids forming complex structures 35. Functions of the individual protein residues in its entire configuration are represented by bulkiness which in turn influences the local conformation of protein 36. In human, the polarity value ranges from 0.403 (983aa) to 45.376 (508aa), hydrophobicity value ranges from -3.822 (507,508aa) to 2.722 (525aa), mutability value ranges from 47.222 (7aa) to 105.444 (238aa), accessibility value ranges from 2.978 (227aa) to 7.878 (604aa), flexibility value ranges from 0.348 (525aa) to 0.513 (277), bulkiness value ranges from 9.090 (598aa) to 19.092 (519aa) (Figure 3). The comparative values of the 6 ProtScale parameters of all the species under study are listed in the Table 2.
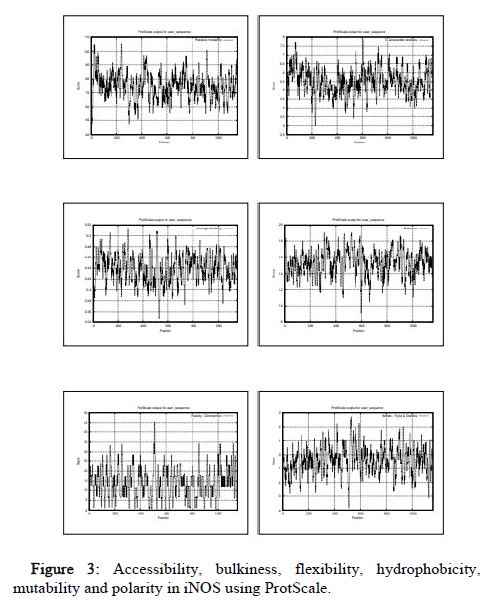
Figure 3: Accessibility, bulkiness, flexibility, hydrophobicity, mutability and polarity in iNOS using ProtScale.
Transmembrane models and signal peptide in NOS2
SignalP 5.0 was used to compute signal peptide in all the organisms under study. It includes scores of Signal Peptide (SP) depending on its types, Cleavage Sites (CS) and others (no signal peptide). Comparing the scores in all the species the values were very less than the threshold value of 0.5. Therefore, no significant score of signal peptide was found indicating no signal peptide (Figure 4).
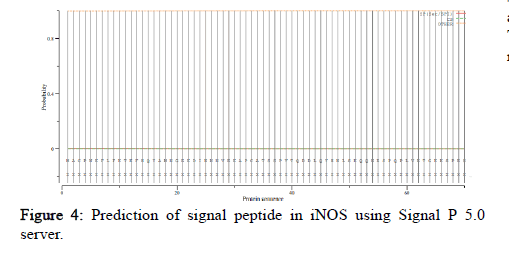
Figure 4: Prediction of signal peptide in iNOS using Signal P 5.0 server.
The TMpred server results show that there are two possible predicted models for transmembrane helixes in NOS2 in most of the species. One of them is strongly preferred model while the other is the alternative model. Prediction score above 500 are taken as significant. The location along with the score values of the strongly preferred model and alternative model of all the species is listed in the Table 3.
Table 3: The comparative values of TMpred parameters in the organisms under study.
Rabbit and bacteria (reductase) sequences do not show any predicted models or score. Horse sequence do not show any predicted alternative model but it contains one strongly preferred model having score 614 (390-410aa) from outside to inside. Gallus shows maximum number of helixes, 5 of them under the strongly preferred model and four of them under the alternative model. Bacteria reductase shows one helix under strongly preferred model with score 890 (365-385aa) from inside to outside and one helix in alternative model with maximum score 531 (360-385aa) from outside to inside. Human shows 5 helixes. The strongly preferred model includes three helixes with scores 1387 (759-779) from outside to inside, 577 (665-683aa) from inside to outside, 590 (554-574aa) from outside to inside. The alternative model includes two helixes with scores 577 (665-683) from inside to outside, 1387 (759-779aa) from outside to inside as shown in Figure 5.
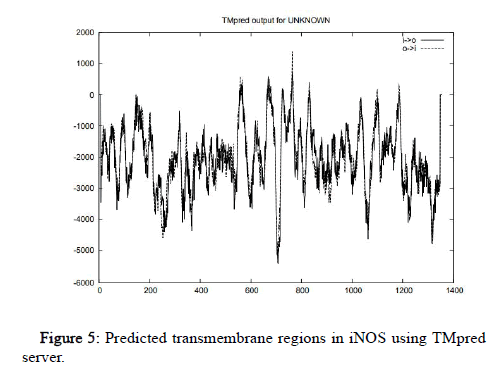
Figure 5: Predicted transmembrane regions in iNOS using TMpred server.
NOS2 is cytoplasmic and soluble
DeepLoc-1.0 was used to calculate the likelihood values of many different sub-cellular localizations of the protein. It also includes the likelihood values of the protein type i.e. soluble or membrane. The likelihood value of human NOS2 for cytoplasm is 0.6854, Golgi apparatus is 0.0899, peroxisome is 0.0681, cell membrane is 0.0474, lysosome/vacuole is 0.046, nucleus is 0.0235, endoplasmic reticulum is 0.0194, extracellular is 0.0105, mitochondrion is 0.0078, plastid is 0.0019, soluble type is 0.8492, membrane type is 0.1508. Similarly, the sub cellular localization of NOS2 in other species was also predicted. The hierarchical tree representation of human NOS2 predicted by DeepLOC-1.0 is shown in Figure 6.
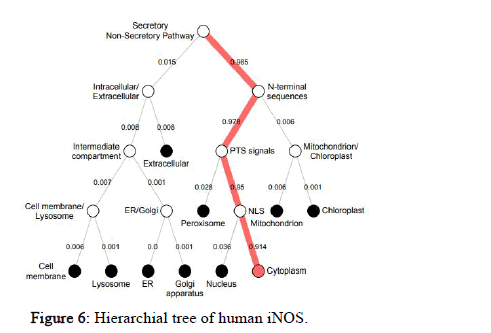
Figure 6: Hierarchial tree of human iNOS.
Prediction of regulatory elements and transcription factor binding sites (TFBS)
The DIRE server was used for identification of distant Regulatory Elements (REs) and transcription factors with their binding sites. The DIRE server is available only for human, mouse and rat. The program was run with default settings of 5000 for the random set of genes. The results of this server do not predict any regulatory elements or transcription factors related to NOS2. Rat contains one regulatory element i.e. promoter and corresponds to 33% of the total regulatory regions in NOS2. It contains only one transcription factor SEF1 at position 238. Mouse contains three regulatory elements i.e. two promoters comprising 67% and one intron comprising 33%. Promoter contains two transcription factors i.e. SEF1 (at 135aa) and COREBINDINGFACTOR (at 184aa). Intron contains 5 transcription factors i.e. DELTAEF1 (0aa), NRSF (150aa), GCM (197aa), NRSF (242aa), NFMUE1 (320aa) (Table 4).
Table 4: Regulatory elements, their type, locus and candidate transcription factors with their positions.
The transcription factors along with their importance and % of occurrence is shown in the Table 5.
Table 5: The list of TFs in iNOS of Rat and Mouse determined by DiRE with their occurrence and importance.
The 33% of occurrence is same for all the transcription factors while in mouse the maximum score (0.33073) of importance is predicted in DELTAEF1, while minimum (0.08333) by core binding factor. In rat SEF1 shows the importance rate of 0.32930.
Predicted deleterious and tolerated SNPs in iNOS
Numerous SNP databases suggest that large portion of SNP is nonsynonymous SNPs (nsSNPs). The deleterious ones may lead to prompt phenotypic outcomes while the tolerant ones keep the protein function despite changes in protein structure. The input was provided in the form of nearly ~ 68 rs IDs which were obtained from the literature survey. Among these 7 (rs3730017, rs16966563, rs28999412, rs28730832, rs1137933, rs1060826, rs2297518) were detected by SIFT. Among these, the SIFT score of rs3730017 was found as 0.022 which is <0.05, so it is predicted as deleterious. The SIFT scores of the rest IDs were Ë?0.05, so these were detected as tolerant (Table 6).
Table 6: The deleterious and tolerated nsSNPs detected by SIFT in the coding sequence of iNOS.
Various post-translational modification (PTMs) sites predicted in NOS2
PTMs regulate numerous cellular processes influencing the protein interaction network while their deregulation might result in modulation of the pathway and finally to the outcome of a disease.
NetPhos is used for the prediction of phosphorylation sites at tyrosine (Y), Serine (S) and Threonine (T) residues in the protein sequences. The proteins are considered as phosphorylated when the score ≥ 0.5. It is seen that maximum number of phosphorylation occurs at serine followed by threonine. Tyrosine shows minimum number of phosphorylation. If observed individually, serine phosphorylation occurs maximum in drosophila and minimum in bacteria (oxygenize). Threonine phosphorylation occur maximum in rat and minimum in bacteria (oxygenize). The tyrosine phosphorylation occurs maximum in drosophila and minimum in alligator.
The graphical representation of various Post Translational Modifications (PTMs) with their maximum and minimum scores in organisms along with the human PTM scores is shown in Figure 7.
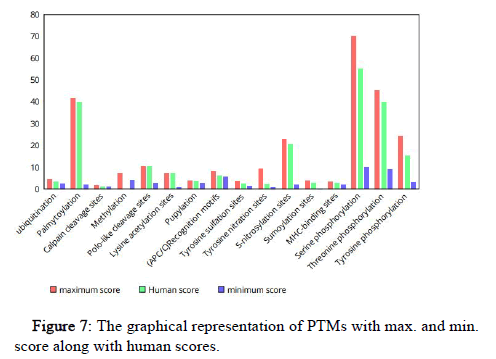
Figure 7: The graphical representation of PTMs with max. and min. score along with human scores.
BDM-PUB serves to predict the sites of ubiquitination in NOS2 using Bayesian Discriminant Method. The ubiquitination is one of the major modifications in a protein which leads to its degradation in eukaryotes playing an important role in DNA repair and cell cycle regulation. Ubiquitination has been shown to occur in the lysine sites of the peptide sequence in all the species. Maximum ubiquitination occurs in Pipra with 4.50 (1143aa) and minimum in bacteria (reductase) with 2.30 (75aa).
CSS-PALM server is used for identifying palmitoylation sites in any sequence. Palmitoylation is the covalent attachment of fatty acids (palmitic acid to cysteine). Palmitoylation increases the membrane affinity and surface hydrophobicity through palmitic acid addition to the protein substrates. It has been shown to occur in the cystine residues of the peptide sequence in all the species. Maximum score is seen in Pipra with 41.74 (3aa) and minimum in bacteria reductase with 1.81 (99aa).
GPS-CCD tool was used for prediction of the calpain-cleavage sites in NOS2. Calpains constitute a very important family of Ca2+dependant cysteine proteases containing a nucleophilic cysteine in the catalytically active sites. General degradation and limited proteolysis are the two modes by which calpain activation takes place. The maximum score is seen in case of horse and monkey with 1.562 (744aa positions) in both. The minimum score was predicted in rat with 0.799 (738aa).
GPS-MSP tool is utilized for identifying methylation of proteins which is considered as an important reversible PTM. The methylation can occur on either side chain or backbone of amino acids residues which includes Arginine (R), Lysine (K), Alanine (A), Proline (P), Asparagines (N) and Histidine (H). Arginine methylation is classified into mono (R.mono), asymmetry di- methylation (R.a.di) and symmetry di-(R.s.di) while Lysine methylation is classified into mono-(K.mono), di-(K.di) and tri-methylation (K.tri) 45. Among R.di type methylation maximum predicted score is of cow with 7.16 (750aa). Along with R.di type of methylation aplysia also shows K.mono type of methylation at 142aa position.
GPS-POLO tool was used for identifying Polo like kinase (Plks) which play crucial roles in orchestrating the cell cycle progression via phosphorylation dependent molecular recognition and phosphorylation. Plks have Polo-Box Domain (PBD) in the C-terminal non catalytic region for proper sub-cellular localization of Plks during multiple stages of mitosis. Among phosphorylation type maximum score predicted is of alligator with 5.721 (862aa) and minimum of cow with 2.667 (663aa). Among Phospho-binding type maximum score predicted is of human with 10.214 (77aa) and minimum of Gallus with 6.661 (651aa). No polo-like cleavage sites has been predicted in bacteria (oxygenize).
GPS-PAIL 2.0 tool predicts the acetylation processes in the proteins which is of two types i.e. Nα-terminal acetylation which is rare in prokaryotes but 85%in eukaryotic proteins while the other one is Nε- lysine acetylation which is less common but one of the most important and ubiquitous PTMs conserved in prokaryotes and eukaryotes. Acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). GPS-PAIL software was used for prediction of substrates and sites of 7 HATs (Histone acetyltransferases) including CREBBP, EP300, HAT1, KAT2A, KAT2B, KAT5, and KAT8. No lysine acetylation sites were predicted in rabbit. Lysine acetylation for human is predicted for 5 HATs i.e. EP300 at 6aa position, KAT2A at 66aa position, KAT2B at 132aa position, KAT5 at 66aa position, KAT8 at 105aa position. Ostrinia shows maximum lysine acetylation sites predicted for 6 HATs. Bacteria (oxygenize) show lysine acetylation at 351aa by CREBBP while bacteria (reductase) show lysine acetylation at 108aa position by KAT2B.
GPS-PUP was used to identify Prokaryotic Ubiquitin like protein (PUP) modification of which is the pupylation. Despite the enzymology of both pupylation and ubiquitylation being different, ubiquitin and PUP has quite similar functions. The maximum score is predicted in danio rerio with 3.803 (501aa) and minimum in case of rabbit with 2.709 (275aa). Bacteria oxygenize and reductase show significant scores with 3.488 (28aa) and 3.354 (143aa). No pupylation site was predicted in alligator.
GPS ARM was used to find the APC/C (Anaphase promoting complex) which is an E3 ubiquitin ligase that marks target cell cycle protein for degradation by 26S proteasome. The D-box and the KENbox are the major APC/C recognition motifs. The destruction box or the D-box follows a minimal consensus of RXXL (where X is any amino acid). No recognition motifs are detected in rat, mouse, bacteria, cow, aplysia, dog, rabbit, and Rhincodon. The maximum score of 8.027 (572-575aa) is predicted in alligator while the minimum score of 5.662 at (752-755aa) position in also predicted in the same species.
GPS-TSP tool is used to identify tyrosine sulfation which is a ubiquitous PTM which modifies proteins in a secretory pathway. The maximum score is seen in Rhincodon with 3.406 (859aa) and minimum in bacteria oxygenase with 1.163 (196aa). Bacteria (reductase) do not show any tyrosine sulfation sites.
GPS YNO2 tool is used to predict the tyrosine nitration of proteins which is a kind of covalent post translational modification and adds a nitro group (-NO2) to onto either one of the two ortho-carbons of the aromatic ring of the tyrosine residues. The maximum score for cutoff 0.554 (Cluster A) is seen in rabbit with 1.915 (2aa). The maximum score for cutoff 1.065 (Cluster B) is seen in pipra with 2.056 (1058aa). The maximum score for cutoff 0.828 (Cluster C) is seen in bacteria reductase with 4.15 (531aa). The maximum score for cutoff 1.16 (Cluster D) is seen in mouse with 9.214 (925aa). Rat and monkey did not show any score in cutoff 1.16 (Cluster D). Bacteria oxygenase and reductase did not show any score in cutoff 1.065 (Cluster B). Scores were obtained for all species in cutoff 0.828 (Cluster C). Human, rat, mouse, bacteria oxygenase, dog, horse, monkey, ostrinia, rhincodon did not show any score in cutoff 0.554 (Cluster A).
GPS-SNO 1.0 tool is used to predict the S-nitrosylation of proteins which includes covalent attachment of a nitrogen monoxide group to the thiol side chain of cysteine. The maximum score for cutoff 1.67 (Cluster A) is seen in Alligator with 2.36 (344aa). The maximum score for cutoff 2.454 (Cluster B) is seen in mouse with 4.532 (110aa). The maximum score for cutoff 20.743 (Cluster C) is seen in Gallus with 22.686 (1066aa). Danio rerio, aplysia, bacteria oxygenase did not show any score with cutoff 1.67 (Cluster A), while Rhincodon, ostrinia, dog, horse, monkey, cow, rat, bacteria oxygenase did not show any score with cutoff 20.743 (Cluster C). Interestingly, no S- nitrosylation sites were predicted in rabbit and bacteria reductase.
GPS-SUMO 2.0 tool is used to predict the small ubiquitin like modifier (SUMO), a 97-residue protein that is attached covalently to lysine residues on the target protein. Sumoylation sites follow a canonical consensus motif φ–K-X-E (where is hydrophobic amino acid and X is any amino acid residue). The maximum scores of cutoff 0.13 (φ-K-X-E) has been seen in Danio rerio with 2.744 (1037aa) and minimum in Gallus with 0.13 (155aa). The maximum scores of cutoff 2.64 (non-consensus) has been seen in drosophila with 3.926 (1122aa) and minimum in bacteria oxygenase with 2.809 (307aa). No sumoylation sites of cutoff 0.13 (φ-K-X-E type) were been seen in Rhincodon, Pipra, Aplysia and Bacteria (oxygenase) while no sumoylation sites of cutoff 2.64 (non-consensus type) were observed in rabbit. Alligator and Bacteria (reductase) did not show any of the sumoylation types.
GPS-MBA 1.0 tool is used to predict MHC class- ll haplotype, l- Ag7.The score is maximum in Danio rerio with 3.279 (3-11aa) and minimum in bacteria oxygenase with 1.797 (102-110aa).
Details for each PTM among all the organisms used in this study are provided in the supplementary material.
NOS2 interactions with other proteins
The STRING 11.0 database provides estimation of protein-protein interactions including both direct as well as indirect ones and the view of STRING network is provided in the Figure 8. It shows maximum interaction with CALM1 (Calmodulin1) followed by TNF (Tumor Necrosis Factor), ARG2 (Arginase2), ARG1 (Arginase1), SRC (SRC Proto-oncogene/Non-Receptor Tyrosine Kinase), CALM3 (Calmodulin 3), CALM2 (Calmodulin 2), CAT (Catalase), JUN (Jun Proto-oncogene/AP-1 Transcription Factor Subunit), and minimum interaction with STAT3 (Signal Transducer and Activator of Transcription 3).
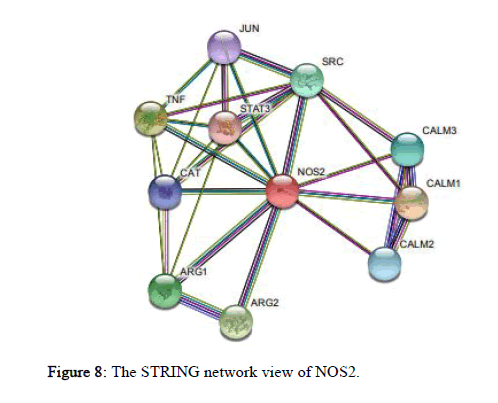
Figure 8: The STRING network view of NOS2.
Discussion
In this study we actually attempted to give a comparative analysis of the structure and function of NOS2 between different species using various bioinformatics tools. Multiple sequence alignment was used to identify important sites such as binding sites, predicting structural and functional characteristics to suggest oligonucleotide primers for PCR and locating the conserved domains. So, it will help in better understanding of protein evolution at the molecular level. While analyzing it we found that amino acid sequences are mostly conserved from aplysia to dog whereas bacteria (oxygenize) shows no similar conserved sequences. Phylogenic profiles constructed after multiple sequence alignment can be used to infer functions of the protein in the entire species. Closely related species are expected to have high similarity in their molecular sequences. From this study it could be inferred that the sequences are conserved throughout the ancestry although they have difference in genomes.
While analyzing the domains we found that the four domains i.e. oxygenize domain, flavodoxin domain, FAD domain, oxidoreductase NAD- binding domain are same in all the species but the difference lies in the position in which these domains are present in different species. This suggests that the function of all the domains is conserved in all the species amid evolution but surprisingly the rabbit showed only the FAD and oxidoreductase NAD binding domains. So, only the reductase domains were found in it. Therefore, it could be inferred that maybe the function of oxygenize domain is performed by any other sequence within iNOS or maybe any other protein. Identification of the protein domains provides view into the functions of that protein.
While investigating the physico-chemical characteristics in NOS2 we found that GRAVY (Grand average of hydropathicity) value is negative in all the species. This indicates that NOS2 is hydrophilic in all the species. So, this may inversely affect the folding of protein, side chain packing and the stability of the protein. The protein stability in test tube is estimated by the instability index (II). The instability index (II) value is >40 in all the species indicating instability but surprisingly bacteria reductase have the value 33.28 indicating its stability. The instability index can provide essential information in the pharmaceutical industry where the expanding utilization of recombinantly expressed protein has highlighted some issues regarding their stability amid long term storage. Aliphatic index value is directly proportional to the thermo stability of the protein. Bacteria reductase with the value of 92.13 is the highest thermostable while aplysia with the value 74.68 is least thermostable. During the pathological conditions there is an alteration in the in vivo half-life. So understanding the protein degradation rates would help in designing drugs related to various human disorders. Overall identifying the physico-chemical properties of NOS2 might help to unravel the physiological functions related to the protein. The structural and biochemical features of NOS2 which include bulkiness, accessibility, mutability, polarity, hydrophilicity and flexibility are known to play an important role for retaining protein structure and stability.
Efficiency of protein secretion is strongly determined by signal peptide. As the value of signal peptide is less than the threshold i.e. 0.5, so no signal peptide is detected in NOS2 in all the species. Proteins without signal peptide may be secreted by unconventional mechanisms and may stay in cytosol for rest of translation. The two possible transmembrane helix models predicted serves in recognition of functional regions in NOS2. While investigating the sub-cellular localization of NOS2 we found that that likelihood value of cytoplasmic location and soluble type are of maximum value in all the species which indicate that NOS2 is cytoplasmic and soluble. The prediction of sub-cellular location is a very important step to understand the protein function.
The information of Transcription Factors (TFs) and Transcription Factor Binding Sites (TFBS) may be useful to unravel different ways to modulate the expression of NOS2. The TFs and TFBS, directly influencing the NOS2 expression could be targeted for achieving desired alterations. Among all the 7 SNPs detected by SIFT, SNP of rs3730017 which is located on exon 7 and associated with asymptomatic malaria was detected as deleterious. This may be explored for studying how it impacts the NOS2 protein function and structure.
Numerous post-translational modifications were predicted among which phosphorylation, methylation, palmitoylation, sumoylation, Snitrosylation, calpain cleavage sites, pupylation, lysine acetylation, and ubiquitination were the major PTM sites. The enzymatic activity of iNOS has been modulated by phosphorylation at multiple sites which may facilitate crosstalk between NO and other signaling pathways via different protein kinases. Phosphorylation of iNOS occurs at multiple residues including serine, threonine, and tyrosine which may be associated with either activation or inactivation of enzyme directly or by modulating other regulatory domains of iNOS. Phosphorylation of iNOS by Src kinases helps in regulation of iNOS and NO production. Hyperactivation of c-Src has been reported in the development of cancer, mainly lung cancer. So, Src inhibition has been investigated as the potential therapy for non- small cell lung cancer in Phase l and ll clinical trials. Ubiquitination of iNOS is required for its degradation. Deregulation of the ubiquitination plays an important role in cancer progression, muscular dystrophy, neurodegenerative disease and autoimmunity. Similarly pupylation also resembles eukaryotic ubiquitination but the difference is that it occurs in Mycobacterium in which it tags protein for degradation via proteosome. Recently PUP (prokaryotic ubiquitin like protein) has been identified as a tagging method in prokaryotes that can be coupled to its targets by deamidation through dop (PUP deamidase/ depupylase) following conjugation which is catalyzed by PafA (PUP protein ligase). Although the details of pup protease system needs further characterization, discovery of degradation mechanism opens the door to investigate the dynamic protein regulation in Mycobacterium which could be targeted by pathogen specific drugs. As expected, Prokaryotes (bacteria) should have shown maximum score for pupylation but predicted results have shown that maximum score is of Danio rerio (zebra fish). During the host defense, mechanism of our body cells controlling the NO synthesis levels may somehow be revealed by degradation of iNOS. The trafficking, membrane- tethering and compartmentalization of various proteins is modulated by palmitoylation. Previous studies have shown that palmitoylation of iNOS at Cys-3 is required for proper intracellular traffic and NO synthesis. Pharmacological inhibition of palmitoyl transferases involved in NOS2 thioacylation might be a useful approach in dealing with inflammatory diseases associated with increased NOS2 levels. Knowing the exact positions of the calpain cleavage sites is very important to understand the working mechanism of calpain as location of cleavage sites are closely related to how calpain precisely modulate gene expression, signal transduction, cell death and apoptosis. Presently, there are 16 known calpain isoform genes in humans. Calpain activation is an early event in cellular response to pathological conditions. This response if sustained may lead to pathological conditions including muscular dystrophy, diabetes and tumorigenesis. Methylation may be a unique molecular mechanism which has been reported in the transcriptional regulation of iNOS gene expression in mensangial cells. Hypermethylation of iNOS gene promoter inhibits its transcription, which may be exploited in the therapeutics. Recent study suggests that acetylation is the key component of histone code. It has been suggested that acetylation of Stat 1 protein leads to decreased Stat 1 binding to the promoter of iNOS. This results in inhibition of IFN-gamma mediated iNOS expression. Acetylation of Stat 1 protein may be exploited therapeutically to down regulate iNOS expression in pro-inflammatory states. The identification of sumoylation sites and SUMO-interaction Motifs (SIMs) is the basis for understanding regulatory mechanisms and biological functions of SUMOs (small ubiquitin like modifiers) which may provide potential targets for further diagnostics and therapeutics. Studies have shown that sumoylation plays an important role in regulating NOS2 expression in astrocytes. Increasing the sumoylation processes can reduce NOS2 expression which may provide novel targets in a variety of neurological diseases. The NO produced by S-nitrosylation of NOS2 reacts with EGFR (epidermal growth factor receptor) and Src- signaling to form a nexus for driving multiple molecular pathways that leads to basal-like breast cancer phenotype in human cell lines. So, NO inhibition may provide a novel strategy for treating basal-like breast cancer. MHC class- ll haplotype, I-Ag7 is strongly linked to susceptibility to T1D (Diabetes mellitus type 1) in the Non-obese Diabetic (NOD) mouse. Identification of the I-Ag7 epitopes is considered important to understand the molecular mechanisms of T1D and then designing the immunotherapeutic peptides. It has been proved that over expression of NOS2 leads to the increase in MHC II cell surface expression in Dendritic Cells (DC) which leads to their degradation. Protection of CD74 (the MHC IIassociated invariant chain) degradation could be one of the mechanisms contributing to the increase in antigen presenting efficiency of maturing dendritic cells, so inhibition of NOS2 expression may somehow reduce the degradation of CD74. Clinical evidence suggests that aberrant expression of polo like kinase (Plk -1) is closely associated with tumorigenesis. Both the kinase domain as well as Polo-Box Domain (PBD) can be considered as attractive targets for designing the anticancer chemotherapeutics. APC/C (Anaphase promoting complex) mediated degradation also helps in regulating Rho GTPase activity, axon growth, cell adhesion and glycolysis. In this regard identifying the APC/C specific degradation substrates is required to understand the regulatory roles and molecular mechanisms of APC/C. Tyrosine sulfation is an important PTM which plays an important role in regulating chemotaxis, inflammatory response and cell adhesion. In animals sulfation is catalyzed by two closely related Tyrosyl Protein Sulphotransferases (TPST- 1and TPST-2). It is convinced that tyrosine nitration of protein plays a critical role in pathology as well as physiology. Many studies have linked nitration to the loss of function, but again the concept of tyrosine nitration as “gain of function” was raised by some reports. Although a very few protein and tyrosine could be nitrated, but still it has very important role in influencing phosphorylation of tyrosine, biomarker for oxidative stress, altering the functions and structure. Current advances have shown the important roles of protein tyrosine in immunomodulation, apoptosis, cell cycle, cell death and ageing.
While we were investigating the protein-protein interactions we found that iNOS shows maximum interaction with CALM1 which is involved in calcium linked pathways. Second-most interacting partner of iNOS is TNF which is linked to tumor Necrosis Factor signaling. ARG2 is involved in metabolism and arginine synthesis. Related pathways of ARG1 include ATF-2 transcription factor network and metabolism. Related pathways of SRC include Thromboxane A2 receptor signaling and Development dopamine D2 receptor transactivation of EGFR. CALM3 is related to genetic pathway which regulates centrosome cycle progression by cytokinensis. CALM2 is a member of calmodulin gene family while CAT is related to Wnt/β- catenin signaling. JUN is related to JNK signaling pathway. Minimum interaction predicted is with STAT3 which is related to JAK/STAT signaling pathway. The involvement of all these protein-protein interactions with iNOS under various disease and physiological conditions and their regulation is still uncertain. Future studies on the iNOS interactions with these proteins may lead to more knowledge about the pathways, mechanisms, and the interlinkage between the different associations of different proteins for targeting the drug ability of the molecule.
Conclusion
The NO generated by iNOS is involved in immunomodulatory, antitumor mechanisms. Dysfunctional induction in expression of iNOS seems to be involved in various human disorders. So, there is a need to tightly regulate the iNOS which includes alteration in expression at both transcriptional as well as post-transcriptional levels. Multiple sequence alignment was done and the evolutionary links of iNOS from 18 different species from arthropods to mammals including bacteria was investigated to understand the protein evolution at the molecular level and to find whether the sequence is conserved. Identification of domains provided insights into functions of iNOS. The comparative analysis of physico-chemical, biochemical and structure features of NOS2 revealed different physiological properties, information about protein stability and structure of the gene. Though no signal peptide was detected but two possible transmembrane helix models were predicted helping in the recognition of functional regions. Through no regulatory elements or Transcription Factors (TF) were detected in human NOS2 but 8 TF were detected in mouse and rat in the promoter and intron regions of NOS2 which may directly influence the NOS2 expression and targeted to achieve desired alterations. The level of NOS2 gene product is altered by polymorphism in the promoter region whereas the activity of NOS2 gene product is altered by polymorphism in the coding region. The insilico analysis of the SNPs obtained from the literature survey predicted 1 deleterious SNP of rs3730017 on exon 7, that might have negative impact on the function of protein and ultimately individual’s inclination to several diseases. Overall, comparative in silico analysis of iNOS unravels various physiological properties, regulatory activities, pathways, and its interaction with different proteins in different species creating a link between them. It is hoped that inhibition of NO by regulating the NOS2 gene will have tremendous impact in multiple disciplines including in the treatment of autoimmune disorders like muscular dystrophy, osteoarthritis and also in tumors.
References
- Xue Q, Yan Y, Zhang R, Xiong H (2018) Regulation of iNOS on immune cells and its role in diseases. Int J Mol Sci 19:3805.
- Cartledge J, Minhas S, Eardley I (2001) The role of nitric oxide in penile erection. Expert Opin Pharmacother 2:95-107.
- Levitzki A (2012) Targeting the immune system to fight cancer using chemical receptor homing vectors carrying polyinosine/cytosine (PolyIC). Front Oncol 2:1-10.
[Googlescholar] [Crossref][Indexed]
- Kröncke KD, Fehsel K, Kolb-Bachofen V (1998) Inducible nitric oxide synthase in human diseases. Clin Exp Immunol 113:147-156.
- Garthwaite J, Boulton CL (1995) Nitric oxide signaling in the central nervous system. Annu Rev Physiol 57:683-706.
- Prast H, Philippu A (2001) Nitric oxide as modulator of neuronal function. Prog Neurobiol 64:51-68.
- Arnold WP, Mittal CK, Katsuki S, Murad F (1977) Nitric oxide activates guanylate cyclase and increases guanosine 3’:5’-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci USA 74:3203-3207.
- Schirripa M, Zhang W, Yang D, Cao S, Okazaki S, et al. NOS2 polymorphisms in prediction of benefit from first-line chemotherapy in metastatic colorectal cancer patients. PLoS One 13:1-16.
- Förstermann U, Sessa WC (2012) Nitric oxide synthases: Regulation and function. Eur Heart J 33:829-837.
- Depre C, Havaux X, Renkin J, Vanoverschelde JLJ, Wijns W (1999) Expression of inducible nitric oxide synthase in human coronary atherosclerotic plaque. Cardiovasc Res 41:465-472.
- Dobutovic B, Sudar E, Tepavcevic S, Djordjevic J, Djordjevic A, et al. (2014) Effects of ghrelin on protein expression of antioxidative enzymes and iNOS in the rat liver. Arch Med Sci 10:806-816.
- Soskic SS (2011) Regulation of Inducible Nitric Oxide Synthase (iNOS) and its Potential Role in Insulin Resistance, Diabetes and Heart Failure. Open Cardiovasc Med J 5:153-163.
[Googlescholar] [Crossref][Indexed]
- Lim MX, Png CW, Tay CY, Teo JD, Jiao H, et al. (2014) Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect Immun 82:4789-4801.
[Googlescholar][Crossref] [Indexed]
- Jones RJ, Jourd’heuil D, Salerno JC, Smith SME, Singer HA (2007) iNOS regulation by calcium/calmodulin-dependent protein kinase II in vascular smooth muscle. Am J Physiol Hear Circ Physiol 292:2634-2642.
- Qidwai T, Jamal F (2010) Inducible Nitric Oxide Synthase (iNOS) gene polymorphism and disease prevalence. Scand J Immunol 72:375-387.
- Hoos MD, Richardson BM, Foster MW, Everhart A, Thompson JW, et al. (2013) Longitudinal study of differential protein expression in an Alzheimer’s mouse model lacking inducible nitric oxide synthase. J Proteome Res 12:4462-4477.
- Casarotto PC, Biojone C, Montezuma K, Cunha FQ, Joca SR, et al. (2018) Inducible nitric oxide synthase (NOS2) knockout mice as a model of trichotillomania. Peer J 6:e4635.
[Googlescholar][Crossref] [Indexed]
- Toulorge D, Schapira AHV, Hajj R (2016) Molecular changes in the postmortem parkinsonian brain. J Neurochem 139:27-58.
- Hu DE, Dyke SOM, Moore AM, Thomsen LL, Brindle KM (2004) Tumor Cell-Derived Nitric Oxide Is Involved in the Immune-Rejection of an Immunogenic Murine Lymphoma. Cancer Res 64:152-161.
- Beere HM (2004) The stress of dying: The role of heat shock proteins in the regulation of apoptosis. J Cell Sci 117:2641-2651.
- Basudhar D, Glynn SA, Greer M, Somasundaram V, No JH, et al. (2017) Coexpression of NOS2 and COX2 accelerates tumor growth and reduces survival in estrogen receptor-negative breast cancer. Proc Natl Acad Sci U S A 114:13030-13035.
- Basudhar D, Bharadwaj G, Somasundaram V, Cheng RY, Ridnour LA, et al. (2019) Understanding the tumour micro-environment communication network from an NOS2/COX2 perspective. Br J Pharmacol 176:155-176.
- Buzzo CD, Medina T, Branco LM, Lage SL, Ferreira LC, et al. (2012) miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc Natl Acad Sci U S A 109:5826-5831.
[Googlescholar] [Crossref][Indexed]
- Buzzo CDL, Medina T, Branco LM, et al. (2017) Epigenetic regulation of nitric oxide synthase 2, inducible (Nos2) by NLRC4 inflammasomes involves PARP1 cleavage. Sci Rep 7:1-12.
- Joseph A, Nair LC, Johnson BS, Thomas PL, Padmanabhan RA, et al. (2019) Transcriptional regulation of NOS2 via STAT5b binding to NOS2 gene promoter mediates nitric oxide production: Relevance in β-cell maintenance. Cell Physiol Biochem 52:141-155.
[Googlescholar] [Crossref][Indexed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi 
