Review Article, J Mol Biol Methods Vol: 1 Issue: 1
Molecular Basis and Engineering of Enzymes Stereospecificity
Aliyu Adamu1,2*, Firdausi Aliyu2, Fahrul Huyop2 and Roswanira Abdul Wahab3
1Faculty of Science, Department of Microbiology, Kaduna State University, Tafawa Balewa way, Kaduna PMB 2339, Nigeria
2Faculty of Biosciences and Medical Engineering, Department of Biotechnology and Medical Engineering, Universiti Teknologi Malaysia, Johor Bahru 81310, Johor, Malaysia
3Faculty of Science, Department of Chemistry, Universiti Teknologi Malaysia, 81310 Johor Bahru, Johor, Malaysia
*Corresponding author : Aliyu Adamu
Faculty of Science, Department of Microbiology, Kaduna State University, Tafawa Balewa way, Kaduna PMB 2339, Nigeria
E-mail: aliyu.adamu.12@aberdeen.ac.uk
Received: January 27, 2018 Accepted: February 15, 2018 Published: February 23, 2018
Citation: Adamu A, Aliyu F, Huyop F, Wahab RA (2018) Molecular Basis and Engineering of Enzymes Stereospecificity. J Mol Biol Methods 1:1.
Abstract
Molecular recognition, particularly enantiomeric specificity of biological molecules is a key consideration in designing drugs, pharmaceutical intermediate and in industrial production of chirally active intermediate. Although the molecular bases of many enzymes stereospecificity are not completely delineated, a number of protein engineering studies were able to enhance and even in some cases invert the stereospecificity of various enzymes. Herein, we review the current understanding on enzymes stereospecificity, and the effects of mutations to the stereospecific pockets due to enzymes engineering to improve stereospecificity.
Keywords: Molecular basis; Stereospecificity; Enzymes; Protein engineering
Introduction
A number of enzymes have the ability to discriminate between enantiomeric substrates or products; such enzymes are referred to as stereospecific/stereoselective enzymes. The substrate specificity of these enzymes are further sub-categorized according to the handedness of the substrates they catalyse. For instance, the L-haloacid dehalogenase from Pseudomonas putida S3 which catalyses the stereospecific hydrolysis of only L-isomer of 2-haloacids [1]. Such enzymes are unique and display chiral preferences in specificity i.e. stereospecificity in their catalysis. The ability of certain microorganisms to produce stereospecific enzyme stems from an evolutionary adaption towards the utilisation of chiral of substrates in their surrounding environment, which are essential for growth [2]. Most compounds in nature are, in fact, chiral. Kinetically, enzyme stereospecificity is expressed as enantiomeric ratio (E), the ratio of specificity constants (Kcat /Km) of the enzyme for the fast (reactive) and slow (non-reactive) enantiomers (Equation 1) [3]. Depending on the enzyme, the fast and slow enantiomers can be D- or L-form of the chiral substrate.
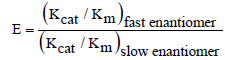 (1)
(1)
Where Kcat the maximum number amount of substrate the enzyme is can convert to product per catalytic site and per unit time. The Km is the Michaelis-Menten constant and it is the substrate concentration when the reaction velocity is half the maximum velocity of the reaction. These constants are mathematically essential to illustrate the chirality of a particular enzyme. An enzyme that is not chiral would typically display an E value = 1, whilst stereospecific enzymes would have E values higher than 1 [3].
Stereospecificity is one of the key properties of enzymes as biocatalysts. Stereospecific enzymes especially hydrolases, are useful in synthesis of pure enantiomers required for pharmaceuticals, agrochemicals and chiral intermediates in chemical industries [4,5]. Although the molecular details of stereospecificity are not completely understood, many studies carried out by different research groups succeeded in enhancing and even in some cases reversing the stereospecificity of various enzymes using protein engineering [6,7]. In this review, we focus on current understanding on enzymes stereospecificity, and success in protein engineering to improve stereospecificity.
Molecular basis for stereospecificity
Determining the molecular basis for stereospecificity of an enzyme requires the identification of the reacting orientations for both fastand slow-reacting substrate enantiomers [3]. The differences between the two orientations along with the accompanying interactions essentially form the molecular basis for the stereospecificity of enzymes. Identifying the reacting orientation for the fast-reacting enantiomer is straightforward; because the reacting atoms of the substrate are oriented in a way that they will best interact with the active site residues, whereas the non-reacting substrate moiety is positioned in the most fitted complementary pocket nearby. Hence, the enantiomeric excess (e.e, %) of the correctly positioned or fast-reacting substrate would in turn, be higher than that of the slow-reacting counterpart. In such cases, the fast reacting substrate enantiomer that an enzyme preferentially catalyses would register a percentage e.e that approaches 100 [8]. Conversely, due to possibilities of compromises, orienting the slow-reacting substrate enantiomer to is often less straightforward. In some instances, certain features in the fast-reacting enantiomer will clash with the slow-reacting enantiomer within the tight active site pocket, as the two enantiomers are mirror images of each other. Therefore the difficulty in identifying the molecular basis of enzyme stereospecificity emanated from orienting the slow-reacting enantiomer [3].
Literature has so far, proposed two approaches that are generally used to orient the slow-reacting enantiomer to interact with the catalytic residues of an enzyme. The first approach involves the fitting of the slow-reacting enantiomer by exchanging two substituents of the fast-reacting enantiomer [9]. The process occurs by swapping the positions of any two of the four substituents that are attached to a chiral atom on the fast-reacting enantiomer to form the slow-reacting orientation, a substrate showing an absolute configuration opposite to that of the fast-reacting substrate. For clarity, absolute configuration defines the spatial arrangement of chiral molecule (group) and its stereochemistry [10]. While the swapping of substituents preserves the location of the stereocenter, it also produces two mismatches between the exchanged substituents and the binding site. It is noteworthy to highlight here that preservation of the stereocenter is important for reactions that involve bond breaking and forming [3] such as dehalogenation.
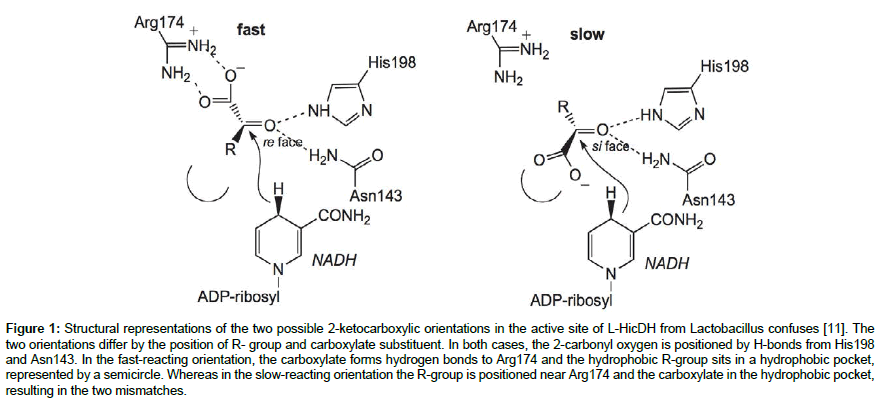
The substituents-exchange concept was observed and demonstrated in the crystal structure of L-2-hydroxyisocaproate dehydrogenase (L-HicDH) from Lactobacillus confuses [11]. X-ray structure of L-HicDH active site reveals that it could fit the substrate, 2-ketocarboxylic acid in two orientations viz. the D- and L-enantiomers (Figure 1). Swapping positions of the R-group and the carboxylate group in either of the substrate orientations generates the opposite orientation of the substrate. This concept of substituentsexchange is the method of choice by many research groups for investigating and improving enzymes stereospecificity. For example, this approach was used to computationally model a slow-reacting enantiomer in order to probe the stereospecificity determinant of an L-2-haloacid dehalogenase from Pseudomonas sp. YL [12].
Figure 1: Structural representations of the two possible 2-ketocarboxylic orientations in the active site of L-HicDH from Lactobacillus confuses [11]. The two orientations differ by the position of R- group and carboxylate substituent. In both cases, the 2-carbonyl oxygen is positioned by H-bonds from His198 and Asn143. In the fast-reacting orientation, the carboxylate forms hydrogen bonds to Arg174 and the hydrophobic R-group sits in a hydrophobic pocket, represented by a semicircle. Whereas in the slow-reacting orientation the R-group is positioned near Arg174 and the carboxylate in the hydrophobic pocket, resulting in the two mismatches.
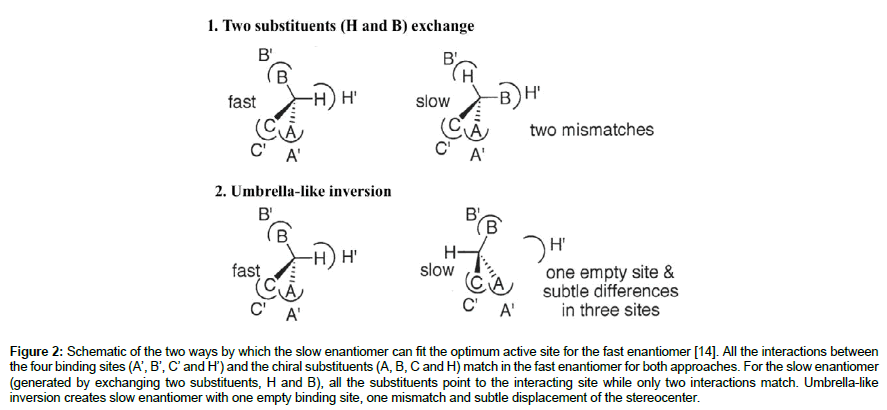
The second approach to orient the slow-reacting enantiomer is via an umbrella-like inversion [13,14]. This approach involves changing the position of a single substituent (usually hydrogen) in the fast-reacting enantiomer to generate the slow-reacting enantiomer orientation. Umbrella-like inversion essentially occur by an inversion through the stereocenter hence, inverting the location of all the four substituents to generate the opposite substrate enantiomer. This is followed by a displacement that reverses the position of the substituents except the hydrogen, relative to their previous positions (Figure 2). Therefore, the hydrogen substituent in the final orientation points to a new direction, and the stereocenter is slightly displaced from its original position. Unlike the substituents exchange approach, umbrella-like inversion generates orientation with lower energy, as the position alteration creates only one mismatch between the substituents and the binding site, rather than two mismatches in the orientation generated by the substituents exchange approach.
Figure 2: Schematic of the two ways by which the slow enantiomer can fit the optimum active site for the fast enantiomer [14]. All the interactions between the four binding sites (A’, B’, C’ and H’) and the chiral substituents (A, B, C and H) match in the fast enantiomer for both approaches. For the slow enantiomer (generated by exchanging two substituents, H and B), all the substituents point to the interacting site while only two interactions match. Umbrella-like inversion creates slow enantiomer with one empty binding site, one mismatch and subtle displacement of the stereocenter.
Re-orientation of a substrate via umbrella-like inversion often occurs in catalytic reactions that are executed adjacent to the stereocenter, and seldom occur in reactions that involve formation and breaking of bond at the stereocenter [3,15]. X-ray structures of enantiomeric substrate configurations bound to various enzymes suggest umbrella-like inversion exist more commonly [14]. For example, comparing the crystal structure of transition state of the fast- and slow-reacting enantiomers of menthol in the active site of Candida rugosa lipase, Cygler and colleagues showed the slowreacting enantiomer is oriented in an umbrella-like orientation [16].
Prediction of stereospecificity
By nature, the active sites of many enzymes are chiral; hence they specifically prefer specific enantiomer of chiral substrates and inhibitors. Therefore, substrate stereopreference needs to be considered when choosing a biocatalyst for reaction that involves specific substrate enantiomer. Based on X-ray crystal structures of enzyme-substrate complex and observed stereospecificity, generalisation was attempted in order to summarize the earlier results and to guide prediction of new substrate behaviours. These generalisations are either based on the substrate properties, in which case are referred to as ‘empirical rules’ [3], or based on the active site properties and are called ‘box models’ [17].
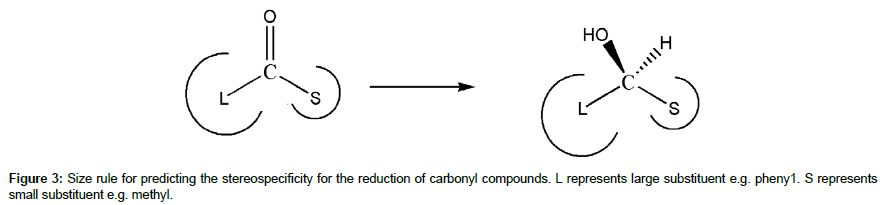
The empirical rule model specifically focuses on the features of the chiral substituents such as shape and size. This is because the strongest destabilising intermolecular interaction involves steric clash between atoms, which is greatly influenced by the size and shape of the substituents. Other rules also consider polar and nonpolar feature of the substituents in predicting the stereospecificity. The simplest empirical rule only specifies the relative size of the chiral substituents (e.g. large, small or medium); hence it is known as size rule [18]. This rule predicts the stereospecificity of reducing ketones based on relative size of the two substituents adjacent to the carbonyl group (Figure 3). The rule suggested that increasing the difference in size of the two substituents could enhance the stereospecificity.
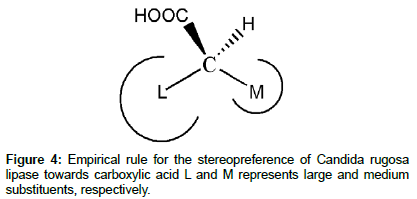
Another example of empirical rule is the one developed by the Kazlauskas group to predict which enantiomer of chiral carboxylic acids preferred by Candida rugosa lipase (Figure 4) [19]. This rule is only reliable for purified Candida rugosa lipase, and it is also based on the size of the chiral substituents. One can interpret this rule as: first, drawing the carboxylic group pointing out from the page towards the reader, and the hydrogen of the substrate pointing inside of the page. Second, imagine a margin along the C−COOH bond that slits the substrate into two parts. The enantiomer with the larger substituent on the left would be the fast-reacting enantiomer.
The advantages of empirical rules in stereospecificity prediction are their straightforwardness, easy application and applicability on a wide range of substrates [3]. However, the rules do not provide detailed information on the molecular basis enzyme stereospecificity. Although several specific substrate rules offer more details, they can only be applicable to specific type of molecule [3].
In order to provide more details regarding stereospecificity of enzyme catalysis, many research groups suggested box models of stereospecificity prediction. These models are based on the active site properties (e.g topology and hydrophobicity), and consider the three- dimensional nature of the molecules [17]. The models were developed using a series of substrates testing to map out the topology and hydrophobicity of the enzymes active site. These series of testing explore the hydrophobicity and the size of the substituents that the active site of the enzyme can accommodate. By so doing, it considers favourable interactions that can occur between the chiral substituents and the binding sites of the enzyme. Hence, the generated models can provide more accurate predictions of the enzyme stereospecificity [17].
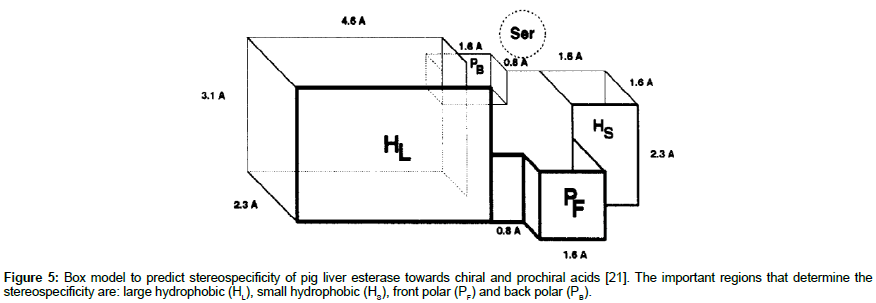
A popular example of box model is for the pig liver esterase. The model was generated to account for the stereospecificity of this hydrolase toward chiral and prochiral acids (Figure 5) [20,21]. To determine the stereospecificity of a substrate, the ester group to be hydrolysed is placed within the serine sphere within the active site. Then, following a set of well-defined rules, the remaining substituents are appropriately fitted into the hydrophobic (H) and polar (P) pockets. After comparing the fit for each enantiomer, the enantiomer that fits better is the preferred enantiomer. However, if both enantiomers fit similarly, then the model predicts low stereospecificity of the tested enzyme.
Figure 5: Box model to predict stereospecificity of pig liver esterase towards chiral and prochiral acids [21]. The important regions that determine the stereospecificity are: large hydrophobic (HL), small hydrophobic (HS), front polar (PF) and back polar (PB).
Alteration of stereospecificity by protein engineering
Although the molecular basis of enantiospecificity is not completely understood, many studies targeted at engineering enzymes to enhance stereospecificity were successful. These successes went beyond increased stereospecificity to even inverting the enzymes stereopreference. An excellent example of such studies is the one reported for a NAD(H)-dependent carbonyl reductase (GoCR) from Gluconobacter oxydans [22]. The crystal structure GoCR and computational models of GoCR-substrate complex were analysed to delineate the molecular basis for the enzyme stereospecificity, and to guide the engineering of the enzyme for substrate preference. Three residues Cys93, Tyr149, and Trp193 in the active site of GoCR were predicted to play a critical role in determining the stereospecificity of GoCR towards ethyl-2-oxo-4-phenylbutyrate. The W193A variant of GoCR generated by site-directed mutagenesis was shown to convert ethyl-2-oxo-4-phenylbutyrate to ethyl (R)-2-hydroxy-4- phenylbutyrate with a significantly improved enantiomeric excess (ee) value of > 99% as compared to only 43.2% for the wild type. The increased stereospecificity is probably attributable to increase in the active site volume due to W193A mutation. Additionally, C93V and Y149A double point mutations were demonstrated to even invert the stereospecificity of GoCR to afford conversion of ethyl-2-oxo-4- phenylbutyrate to ethyl (S)-2-hydroxy-4-phenylbutyrate with ee of 79.8% [22].
It is important to note that while an increase in stereospecificity can be achieved by using point mutation, inversion of substrate preference involves multiple substitutions of amino acid residues [3]. In most cases, the molecular basis of substrate stereospecificity inversion involves either exchange in positions of two substituents or switch in the location of the catalytic residues. More examples of studies that succeeded in engineering the stereospecificity by rational design and/or directed evolution are listed in (Table 1) [23-30].
| Enzymes | Mutation | Function | ΔE/Δenantiomer | Reference |
|---|---|---|---|---|
| Pseudomonas aeruginosa lipase | L162G | Increase stereospecificity | 1.1 to 34 (p-nitrophenyl 2-methyldecanoate) | [23] |
| Burkholderia cepacia lipase | I287F, A287F | Increase stereospecificity | 44 to 123, 5 to 123 (2-cyclohexyl ethanol) | [6] |
| Agrobacterium radiobacter AD1 halohydrin dehalogenase | W249F | Increase stereospecificity | 150 to 900 (p-nitro-2-brom-1-phenylethanol) | [24] |
| Candida antarctica lipase B | S47A | Increase stereospecificity | 6.5 to 12.5 (1-bromo-2-octanol) | [25] |
| Agrobacterium radiobacter AD1 epoxide hydrolase | I219F | Increase stereospecificity | 17 to 91 (styrene oxide) | [26] |
| Organophosphorus hydrolase | H257Y, F132G, S308G I106G | Stereospecificity reversal | E = 21 (SP)to E > 100 (RP) | [27] |
| Burkholderia cepacia lipase | L17P,P119L L167G L266V | Stereospecificity reversal | E = 33 (S) to E = 38 (R) | [28] |
| Burkholderia gladioliesterase | L135P, I152N V351S H253P | Stereospecificity reversal | E =6.1(S) to E =29(R) | [29] |
| Bacillus subtilis esterase | D188W M193C | Stereospecificity reversal | E >100(R) to E =64(S) | [7] |
| Aryl malonate decarboxylase | G74C C188S | Stereospecificity reversal | E >100(S) to E =32(R) | [30] |
Table 1: Mutations that increase/reverse stereospecificity.
Effect of Mutations to the Stereospecificity Pocket
Mutations closer to the stereocenter or active site of enzymes influence stereospecificity more than mutations far away from the active site [31]. Although the molecular details of enzyme stereospecificity varies among different enzymes, mutations of active site residues generally result to one or more of the following effects to the stereospecificity pocket:
Alteration to size and shape of the binding pocket
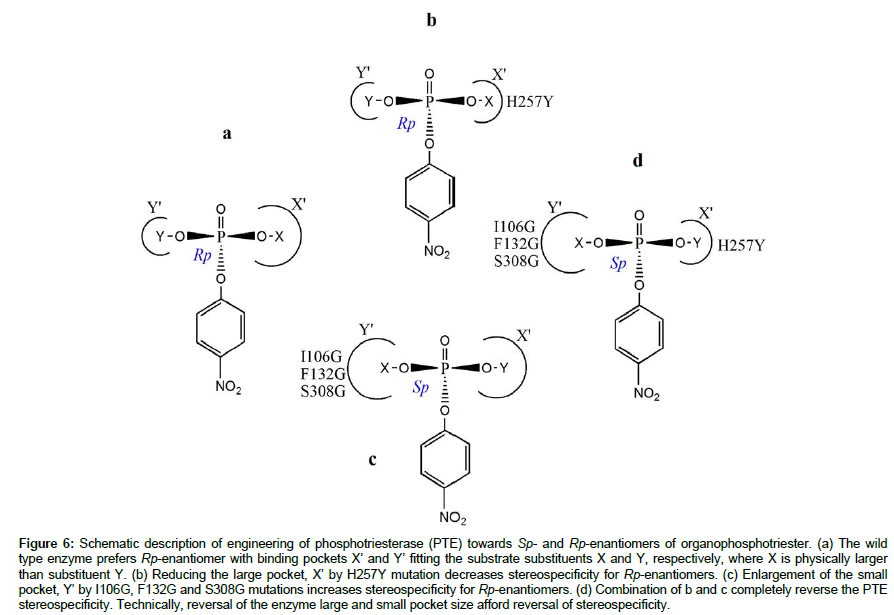
This effect is best described in a study that demonstrated the stereospecificity determinants of phosphotriesterase (PTE) toward organophosphotriester containing phosphorus chiral center connected to various combinations methyl, ethyl, isopropyl and phenyl substituents [32]. The wild type PTE naturally prefers Sp-substrate enantiomers to Rp-enantiomers by factor > 10. Different variants of PTE were generated by rational evolution of the active site residues. Reduction in size of small binding pocket by G60A mutation increased the enzyme stereospecificity toward Sp-enantiomers, while it enlargement by I106G, F132G and S308G mutations significantly increased the specificity constants for Rp-enantiomers by up to 2700- fold, but had little effect on the specificity constants for Sp-enantiomers (Figure 6). Also H257Y variant with reduced large pocket had decreased specificity constant for Sp-enantiomers, whereas those for Rp-enantiomers were unchanged. More interestingly, simultaneous alterations to the size of the large and small binding pockets by the aforementioned mutations resulted in complete reversal of PTE chiral stereospecificity. The reason is that substitution of amino acid residue with large side chain such as tryptophan, phenylalanine and histidine, with residue that has smaller side chain like alanine and glycine would significantly increase the size of the binding pocket such that it fit larger substituents [33]. Conversely, substitution of small residue with a bigger one would reduce the size of the binding pocket, as it will occupy more space within the pocket; hence limit the binding to only small substituents. For example, Marton et al., demonstrated that alteration to the shape and size of the active site entrance of Candida antarctica lipase B due to single point substitution of large with smaller or small with larger residue, I189A, L278V or A282L confers significant modification to the enzyme stereospecificity.
Figure 6: Schematic description of engineering of phosphotriesterase (PTE) towards Sp- and Rp-enantiomers of organophosphotriester. (a) The wild type enzyme prefers Rp-enantiomer with binding pockets X’ and Y’ fitting the substrate substituents X and Y, respectively, where X is physically larger than substituent Y. (b) Reducing the large pocket, X’ by H257Y mutation decreases stereospecificity for Rp-enantiomers. (c) Enlargement of the small pocket, Y’ by I106G, F132G and S308G mutations increases stereospecificity for Rp-enantiomers. (d) Combination of b and c completely reverse the PTE stereospecificity. Technically, reversal of the enzyme large and small pocket size afford reversal of stereospecificity.
Alteration to charge of the binding pocket
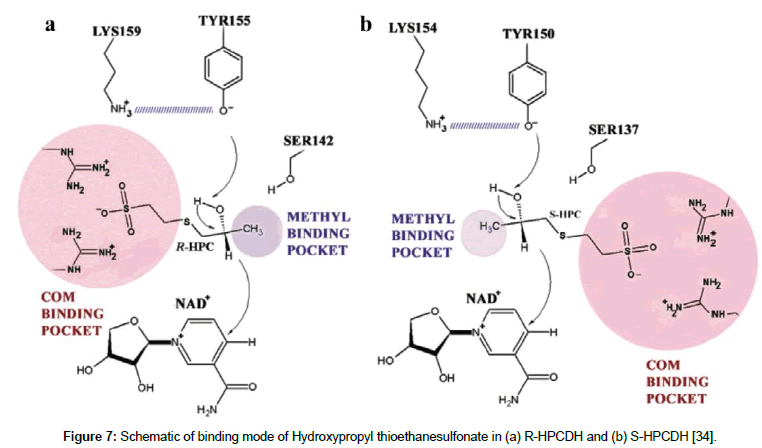
Mutation in stereospecificity pocket that involve electrically charged residues, aspartate, glutamate, arginine, lysine and histidine may cause change in the charge of the pocket. This change alters the integrations particularly electrostatic interactions between the substrate substituents and the binding residues, which in turn would affect the overall binging mode/orientation of the substrate within the pocket. For example R- and S-hydroxypropyl-coenzyme M dehydrogenases (R-HPCDH and S-HPCDH) from Xanthobacter autotrophicus Py2, catalyse the conversion of R- and S- enantiomers of epoxypropane, respectively to the corresponding enantiomers of 2-ketopropyl-CoM [34]. The fundamental molecular rational of each of the R-HPCDH and S-HPCDH catalysis are the electrostatic interactions between two positively charged arginine residues in the CoM binding pocket and the negatively charged oxygen atoms of the sulfonate substituent of the substrate (Figure 7). These electrostatic interactions are responsible for orienting the substrate into it correct orientation within the active site. Although the these enzymes have not been rationally engineered previously, altering the surface charge of the CoM binding pocket by replacing either of or both arginine residues within the pocket with oppositely charged or electrically neutral residues, the activity and/or stereospecificity of the enzymes may significantly be affected.
Figure 7: Schematic of binding mode of Hydroxypropyl thioethanesulfonate in (a) R-HPCDH and (b) S-HPCDH [34].
Steric and stereoelectronic clash
This is when two or more atoms or chemical groups come into proximity of one another particularly within van der Waals radii, and exert mutual repulsion effects [10]. Although the actual amount of energy associated with steric effects in enzyme catalysis was found to be small [35], it can substantially affect the enzymatic reaction. Steric effect, which was bring about by mutating the substrate binding site residues in Pseudomonas fluorescens esterase (PFE) was reported to increase the enzyme stereospecificity towards methyl 3-bromo-2- methylpropionate [36]. The study revealed W28L mutation increase the enzyme stereospecificity relative to the wild type. The explanation is that the W28L mutation brings about substitution of polar tryptophan with non-polar leucine, which removes the electronic repulsive interaction due to steric clash between the electropositive indole ring of the Try28 and the methyl partially induced electropositive moiety of bromomethyl substituent of the substrate. Thus this favours the enantiomer whose bromomethyl groups lies nearest to the position 28.
Hydrophobicity of the pocket
In aqueous environment, water molecules exclude non-polar groups such that they are force to associate with each other. Such property of non-polar groups is referred to as hydrophobicity [10] and has great effect on enzymes activity as it is associated with eight of the twenty amino acid residues (Gla, Ala, Val, Leu, Iso, Met, Phe and Pro) constituting every protein. A protein engineering study of Candida antarctica lipase B (CALB), generated two single point mutants T42V and S47A by site-directed mutagenesis [37]. Both single point mutations, which are situated in the stereospecificity pocket, doubled the stereospecificity of the CALB mutants towards R-enantiomers of secondary alcohols as compered to the wild type. In this case the increased stereospecificity is not attributable to the size effects of the stereospecificity pocket, because threonine and valine are both are have the same number of atoms and the same number and configuration of valence electrons (i.e. isosteric), and serine and alanine have similar molecular volume. Hence, T42V and S47A mutation would not significantly result in major change in the size f the stereospecificity pocket. However, being Thr and Ser hydrophilic and Val and Ala hydrophobic, T42V and S47A mutations increase the hydrophobicity of the stereospecificity pocket. This modifies the hydrogen bond network between residues with the vicinity of the stereospecificity pocket, which in turn perturb the mobility of the residues as well as the substrate moiety placed within the pocket. Thus, these effects cumulatively cause the increase in E value of the single point mutants as compared to the wild type.
Hydrogen bond and other non-bonded interactions
Mutations of the amino acid residues lining the stereospecificity pocket do not only alter the physicochemical properties of the pocket (e.g. size, shape and hydrophobicity), they also change the interactions that occur between the pocket residues and the chiral substituents of the substrate. Hydrogen bond and other non-bonded interactions such as van der Waals and electrostatic interactions are the major interactions that exist between enzyme and substrates. Individual energetic contributions of these interactions are relatively small, but collectively they fuel enzymatic reactions [38,39]. Hydrogen bond interaction between an active site residue of a fluoroacetate dehalogenase and fluorine atom was shown to be a critical requirement for fluoroacetate dehalogenation [40,41]. Mutation of binding site residue in a haloacid dehalogenase, L-DEX from Pseudomonas sp. YL that afford hydrogen bond with the chiral substrate significantly alters the enzyme stereospecificity [12]. L-DEX is absolutely stereospecific towards L-enantiomers of haloacids. Site-directed mutation of nonpolar phenylalanine positioned in the stereospecificity pocket to polar tyrosine (F60Y) created a mutant with activity towards D-enantiomers of haloacids. The molecular basis for the gain of activity toward D-enantiomers of haloacids in the mutant enzyme is the introduction hydrogen bond interaction due F60Y replacement, which is absent in the wild type.
References
- Thasif S, Hamdan S, Huyop F (2009) Degredation of D,L-2-chloropropionic acid by bacterial dehalogenases that shows stereospecificity and its partial enzymatic characteristics. Biotechnology 8: 264-269.
- Nandi N (2009) Chiral discrimination in the confined environment of biological nanospace: reactions and interactions involving amino acids and peptides. Int Rev Phys Chem 28: 111-167.
- Kazlauskas R (2008) Engineering enantioselectivity in enzyme-catalyzed reactions. Protein Engineering Handbook 1, 2: 15-46.
- Patel RN (1997) Stereoselective biotransformations in synthesis of some pharmaceutical intermediates. Adv Appl Microbiol 43: 91-140.
- Singh R, Kumar M, Mittal A, Mehta PK (2016) Microbial enzymes: industrial progress in 21st century. 3 Biotech 6: 174.
- Ema T, Fujii T, Ozaki M, Korenaga T, Sakai T (2005) Rational control of enantioselectivity of lipase by site-directed mutagenesis based on the mechanism. Chem Commun (37): 4650-4651.
- Bartsch S, Kourist R, Bornscheuer UT (2008) Complete inversion of enantioselectivity towards acetylated tertiary alcohols by a double mutant of a Bacillus subtilis esterase. Angew Chem Int Ed Engl 47: 1508-1511.
- Finn MG (2002) Emerging methods for the rapid determination of enantiomeric excess. Chirality 14: 534-540.
- Mugford PF, Wagner UG, Jiang Y, Faber K, Kazlauskas RJ (2008) Enantiocomplementary enzymes: classification, molecular basis for their enantiopreference, and prospects for mirror-image biotransformations. Angew Chem Int Ed Engl 47: 8782-8793.
- Inczedy J, Lengyel T, Ure A (1997) International Union of Pure and Applied Chemistry, Compendium of Analytical Nomenclature, definitive rules. Blackwell Science, 474,699.
- Niefind K, Hecht H-J, Dietmar (1995) Crystal structure of l-2-hydroxyisocaproate dehydrogenase from lactobacillus confususat 2.2 Å resolution. An example of strong asymmetry between subunits. J Mol Biol 251: 256-281.
- Kondo H, Fujimoto KJ, Tanaka S, Deki H, Nakamura T (2015) Theoretical prediction and experimental verification on enantioselectivity of haloacid dehalogenase L-DEX YL with chloropropionate. Chem Phys Lett 623: 101-107.
- Colton IJ, Yin DT, Grochulski P, Kazlauskas RJ (2011) Molecular basis of chiral acid recognition by candida rugosa lipase: x-ray structure of transition state analog and modeling of the hydrolysis of methyl 2-methoxy-2-phenylacetate. Adv Synth Catal 353: 2529-2544.
- Mezzetti A, Schrag JD, Cheong CS, Kazlauskas RJ (2005) Mirror-image packing in enantiomer discrimination molecular basis for the enantioselectivity of B.cepacia lipase toward 2-methyl-3-phenyl-1-propanol. Chem Biol 12: 427-437.
- Molecular Basis for the Enantio RJ, Ha H-J (2014) Molecular basis for the enantio- and diastereoselectivity of burkholderia cepacialipase toward γ-butyrolactone primary alcohols. Adv Synth Catal 356: 3585-3599.
- Cygler M, Grochulski P, Kazlauskas RJ, Schrag JD, Bouthillier F, et al. (1994) A structural basis for the chiral preferences of lipases. J Am Chem Soc 116: 3180-3186.
- Weissfloch ANE, Kazlauskas RJ (2001) Choosing hydrolases for enantioselective reactions involving alcohols using empirical rules. In: enzymes in nonaqueous solvents. Methods in Biotechnology, Humana Press, New York, USA.
- Prelog V (1964) Specification of the stereospecificity of some oxido-reductases by diamond lattice sections. Pure Appl Chem 9: 119-130.
- Ahmed SN, Kazlauskas RJ, Morinville AH, Grochulski P, Schrag JD, et al. (1994) Enantioselectivity of candida rugosa lipase toward carboxylic acids: a predictive rule from substrate mapping and x-ray crystallography. Biocatal Biotransformation 9: 209-225.
- Toone EJ, Werth MJ, Jones JB (1990) Enzymes in organic synthesis. 47. Active-site model for interpreting and predicting the specificity of pig liver esterase. J Am Chem Soc 112: 4946-4952.
- Provencher L, Jones JB (1994) A concluding specification of the dimensions of the active site model of pig liver esterase. J Org Chem 59: 2729-2732.
- Deng J, Yao Z, Chen K, Yuan YA, Lin J, et al. (2016) Towards the computational design and engineering of enzyme enantioselectivity: A case study by a carbonyl reductase from Gluconobacter oxydans. J Biotechnol 217: 31-40.
- Reetz MT, Wilensek S, Zha D, Jaeger KE (2001) Directed evolution of an enantioselective enzyme through combinatorial multiple-cassette mutagenesis. Angew Chem Int Ed Engl 40: 3589-3591.
- Tang L, van Merode AEJ, Lutje Spelberg JH, Fraaije MW, Janssen DB (2003) Steady-State kinetics and tryptophan fluorescence properties of halohydrin dehalogenase from agrobacterium radiobacter. Roles of W139 and W249 in the active site and halide-induced conformational change. Biochemistry 42: 14057-14065.
- Rotticci D, Rotticci-Mulder JC, Denman S, Norin T, Hult K (2001) Improved enantioselectivity of a lipase by rational protein engineering. Chembiochem 2: 766-770.
- Rui L, Cao L, Chen W, Reardon KF, Wood TK (2005) Protein engineering of epoxide hydrolase from Agrobacterium radiobacter AD1 for enhanced activity and enantioselective production of (R)-1-phenylethane-1,2-diol. Appl Environ Microbiol 71: 3995-4003.
- van Loo B, Spelberg JHL, Kingma J, Sonke T, Wubbolts MG, et al. (2004) Directed evolution of epoxide hydrolase from A. radiobacter toward higher enantioselectivity by error-prone PCR and DNA shuffling. Chem Biol 11: 981-990.
- Koga Y, Kato K, Nakano H, Yamane T (2003) Inverting enantioselectivity of Burkholderia cepacia KWI-56 lipase by combinatorial mutation and high-throughput screening using single-molecule PCR and in vitro expression. J Mol Biol 331: 585-592.
- Ivancic M, Valinger G, Gruber K, Schwab H (2007) Inverting enantioselectivity of Burkholderia gladioli esterase EstB by directed and designed evolution. J Biotechnol 129: 109-122.
- Terao Y, Ijima Y, Miyamoto K, Ohta H (2007) Inversion of enantioselectivity of arylmalonate decarboxylase via site-directed mutation based on the proposed reaction mechanism. J Mol Catal B Enzym 45: 15-20.
- Morley KL, Kazlauskas RJ (2005) Improving enzyme properties: when are closer mutations better? Trends Biotechnol 23: 231-237.
- Chen-Goodspeed M, Sogorb MA, Wu F, Raushel FM (2001) Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues. Biochemistry 40: 1332-1339.
- Bartlett GJ, Porter CT, Borkakoti N, Thornton JM (2002) Analysis of catalytic residues in enzyme active sites. J Mol Biol 324: 105-121.
- Krishnakumar AM, Nocek BP, Clark DD, Ensign SA, Peters JW (2006) Structural basis for stereoselectivity in the (R) - and (S)-hydroxypropylthioethanesulfonate dehydrogenases. Biochemistry 45: 8831-8840.
- Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, et al. (2006) Electrostatic basis for enzyme catalysis. Chem Rev 106: 3210-3235.
- Park S, Morley KL, Horsman GP, Holmquist M, Hult K, et al. (2005) Focusing mutations into the P. fluorescens esterase binding site increases enantioselectivity more effectively than distant mutations. Chem Biol 12: 45-54.
- Marton Z, Léonard NV, Syrén PO, Bauer C, Lamare S, et al. (2010) Mutations in the stereospecificity pocket and at the entrance of the active site of Candida antarctica lipase B enhancing enzyme enantioselectivity. J Mol Catal B Enzym 65: 11-17.
- Bohác M, Nagata Y, Prokop Z, Prokop M, Monincová M, et al. (2002) Halide-stabilizing residues of haloalkane dehalogenases studied by quantum mechanic calculations and site-directed mutagenesis. Biochemistry 41: 14272-14280.
- Shurki A, Štrajbl M, Villa J, Warshel A (2002) How much do enzymes really gain by restraining their reacting fragments? J Am Chem Soc 124: 4097-4107.
- Nakayama T, Kamachi T, Jitsumori K, Omi R, Hirotsu K, et al. (2012) Substrate specificity of fluoroacetate dehalogenase: an insight from crystallographic analysis, fluorescence spectroscopy, and theoretical computations. Chem Eur J 18: 8392-8402.
- Kamachi T, Nakayama T, Shitamichi O, Jitsumori K, Kurihara T, et al. (2009) The catalytic mechanism of fluoroacetate dehalogenase: a computational exploration of biological dehalogenation. Chem Eur J 15: 7394-7403.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi