Research Article, J Appl Bioinforma Comput Biol Vol: 7 Issue: 3
Profiling of Differential Expression of Genes in Mice Carrying Both Mutant Presenilin 1 and Amyloid Precursor Protein Transgenes with or without Knockout of Î’2 Adrenergic Receptor Gene
Yuan Zhou1,2,3,#, Lintao Chen1,4,#, Xi Zhou1,4, Yechun Pei1,4, Shuangshuang Wei1,4, Anum Mehmood1,4, Yang K Xiang2,5* and Dayong Wang1,2,4*
1Laboratory of Biotechnology and Molecular Pharmacology, Hainan Key Laboratory of Sustainable Utilization of Tropical Bioresources, Hainan University, Haikou, Hainan 570208, China
2Department of Molecular and Integrative Physiology, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA
3Amber Glen Alzheimer’s Association, 1704 Amber Ln, Urbana, IL 61802, USA
4Collage of Biology, Institute of Tropical Agriculture and Forestry, Hainan University, Haikou, Hainan 570208, China
5Department of Pharmacology, University of California, Davis, CA95616, USA
#The first two authors contributed equally to the study
*Corresponding Authors : Dayong Wang
Professor of Biochemistry and Molecular Biology, Laboratory of Biotechnology and Molecular Pharmacology, Hainan Key Laboratory of Sustainable Utilization of Tropical Resource
Hainan University, 817 Nong-Ke Lou, 58 People’s Road, Meilan District, Haikou, Hainan 570208, China
Tel: +86-18789556728/+1-217-721-9757
E-mail: wangdy@hainu.edu.cn
Yang K. Xiang
Department of Pharmacology, University of California, Davis, CA 95616, USA
Tel: 530-752-6895
E-mail: ykxiang@ucdavis.edu
Received: June 05, 2018 Accepted: September 11, 2018 Published: September 18, 2018
Citation: Zhou Y, Chen L, Zhou X, Pei Y, Wei S, et al. (2018) Profiling of Differential Expression of Genes in Mice Carrying Both Mutant Presenilin 1 and Amyloid Precursor Protein Transgenes with or without Knockout of Β2 Adrenergic Receptor Gene. J Appl Bioinforma Comput Biol 7:3. doi: 10.4172/2329-9533.1000155
Abstract
Alzheimer’s disease (AD) is a lifelong progressive neurodegenerative disease related with accumulation of amyloid β peptide (Aβ) produced by processing of amyloid precursor protein (APP) in the brain. In spite of several-decades effort on AD, there is still no medicine used to intervene with its pathological processes. Our previous studies made in transgenic animal models harboring familial AD genes of mutant presenilin 1 and amyloid precursor protein (APP) showed that β2AR gene knock-out (β2AR-KO) is beneficial in senile AD animals. Consistently, an epidemiological study lasted for two decades showed that the sole usage of β blockers as antihypertensive medicines is associated with fewer brain lesions and less brain shrinkage seen in senile AD patients. In order to understand why senile β2AR-KO AD mice had better learning and memory, genomic effects of β2AR-KO in the double transgenic AD mice were investigated. In the analysis, major genomic significance of β2AR-KO was directed to influence proteinprocessing and presentation involving membrane structure and MHC class I and II protein complex, and lysosome and hydrolase activity for protein degradation, which are critical for accumulation of amyloid β peptide, the hallmark of AD.
Keywords: β2 adrenergic receptor; Alzheimer’s disease; Genome; Differential expression; Protein processing and presentation; Lysosome
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease characterized by accumulation of plaques composed of amyloid β peptides (Aβ) in the brain related with abnormal processing and presentation of Aβ by cells [1]. Studies have shown that the number of β2 adrenergic receptor (β2AR) is increased in the prefrontal cortex and the hippocampus of AD subjects, while it is not correlated with age [2]. Since the cortex and the hippocampus are major brain regions that are highly degenerated in AD and the cortex is the earliest to form Aβ plaques, it is supposed that β2AR may play an important role in AD [3]. A study has shown that chronic treatment with β2AR agonist enhances γ-secretase activity, and related Aβ production and formation of Aβ plaques [4]. Using AD transgenic mouse model harboring human familial AD genes of mutant presenilin 1 (PS1) and amyloid precursor protein (APP), we have found that β2AR gene knock-out (KO) improved learning and memory in 1-year-old senile AD mice, although there was a weak decreasing tendency of the performance in 6-month-old young wild type (WT) mice [5]. In addition, removing the gene encoding β2AR increases the survival rate in P301S mutant tau-transgenic mice [6]. An epidemiological study showed that variations in β2AR gene are involved in the pathogenesis of sporadic late onset Alzheimer’s disease (LOAD) [2]. In an epidemiological study made in 2,197 participants conducted from 1991 to 2010, it was found that clinical use of β-blockers was associated with a lower risk of cognitive impairment, and the association was more obvious in senile men who were more than 75-years old [7]. An autopsy study made on 774 brains of male AD patients after death showed that the patients who took β-blockers had less brain lesions and shrinkage than those who took other medications for blood hypertension and those untreated, and their brains showed that they suffered less microinfarcts [8]. A parallel study showed that patients who took β-blockers experienced less cognitive decline as they aged compared to control groups [9].
β2AR which consists of seven transmembrane α-helices belongs to G protein-coupled receptor superfamily and transduces signal via Gαs and also Gβγ proteins [10]. β2AR is distinct from β1AR in that β2AR internalizes after binding to isoproterenol or Aβ [11]. In response to β2AR endogenous ligands norepinephrine and epinephrine, Gαs dissociates from Gβγ to stimulate adenylyl cyclase to produce cAMP, which activates protein kinase A (PKA) and the exchange protein activated by cAMP (Epac) [10,12]. Gβγ dimer interacts with many different proteins, and different combinations of Gβ and Gγ subtypes transduct signals diversly with or without the association of Gα subunit to inhibit or activate various downstream signaling components, including ion channels, G protein-coupled receptor kinases (GRKs), phosphoinositide 3-kinase (PI3K), mitogenactivated protein kinases (MAPK) signaling mediated by small GTPase Ras, phospholipase A and C, and etc [13-16]. The signal transductions mediated by Gαs and Gβγ regulate a few of transcription factors, such as cAMP response element binding protein (CREB), extracellular signal-regulated kinases (ERKs) and etc., therefore, β2AR activation regulates expression of genes, however integrative influence of β2AR on genomic expression is still unknown. β2ARs are expressed throughout the brain, abundantly in the cortex and the hippocampus, which are the two brain regions essential for higher cognitive functions [6]. Despite being involved in fundamental biological processes, β2AR geneknockout (KO) mice are viable and fertile [6].
In the study, a genetic approach was adopted to remove β2AR gene from a double transgenic mouse model of AD that overexpresses mutant human PS1 gene harboring M146L and L286V familial AD mutations, and APP gene (695) harboring Swedish (K670N, M671L), Florida (I716V) and London (V717I) mutations, and the effects of β2AR-KO on genomic expression profiles in PS1/APP transgenic AD mice was investigated.
Materials and Methods
Animals
PS1/APP double transgenic and β2AR-KO/PS1/APP mice in B6 background were described previously [11,17,18], and 1-year old mice were used for analysis of whole genome gene expression. The PS1/APP mice, which are transgenic animal models of Alzheimer’s disease, were purchased from Jackson laboratory (stock number: 006554), which overexpress human PS1 gene harboring two familial AD mutations, M146L and L286V and APP (695) gene with Swedish (K670N, M671L), Florida (I716V) and London (V717I) familial AD mutations [18]. PS1/APP mice were crossbred with β2AR-KO mice to produce β2AR-KO/PS1/APP mice. All animal experimental procedures were approved by the Animal Care and Use Committee of University of Illinois and Hainan University.
Whole-genome expression analysis
One-year-old PS1/APP and β2AR-KO/PS1/APP mice were sacrificed, six mice in each group. The cerebrums were dissected out, and one side of the cerebral hemispheres of each mouse was used for the study. The cerebral hemispheres were homogenized separately on ice, and total RNA was extracted using RNeasy® Lipid Tissue Kit (Qiagen). Whole-genome expression profiles of PS1/ APP Alzheimer’s disease mice and β2AR-KO/PS1/APP mice were tested using the Mouse WG-6 v2.0 BeadChips and HiScan System (Illumina). The Mouse WG-6 v2.0 BeadChips have probes for the NCBI Mouse Reference Sequence (RefSeq) Release 22, including 26,766 manually annotated and reviewed coding transcripts designated as known mRNAs which begin with NM, 6,856 predicted coding transcripts (XM), and 56 annotation-well-established noncoding transcripts (XR). The BeadChips also have probes for 5,659 transcripts described in the Japan RIKEN Functional Annotation of Mouse (FANTOM) 24-6 database, 3,573 additional sequences listed in the RefSeq Release 5 (Build 33.1), and 2,371 sequences listed in the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) set. In the single factor experimental design, PS1/APP transgenic mice were used as background controls for the gene expression profile of the Alzheimer’s disease model, and β2AR-KO was the only factor of treatment.
Results
Principal components analysis for all biological samples
Principal component analysis (PCA) is a multivariate statistical method to analyze internal correlation or variation among original variables by constructing linear combinations of the variables, which were gene expression levels in the study. During the process the dimensionality of original variables is reduced and the newly constructed variables designed to preserve much original information are independent to each other. The linear combination with biggest variance is treated as the first or the primary principal component (PC), which was designated as PC1 in the study, and the second one was PC2. The PCs which were orthogonal to each other effectively show variation of gene expression levels among thousands of noise in each subject. In the study, PC1 showed that β2AR-KO-induced significant change in genomic expression, in that the two experimental groups which are PS1/APP double transgenic AD mice (PA) and β2AR-KO/PS1/APP mice (B2P) were distributed in two separated regions along the PC1 axis; PC1s for PA samples fell into the region from 19 to 39, however, B2P samples were in the region from -40 to -12 (data not shown). In other words, β2AR-KO in PS1/APP mice significantly changed the distribution of PC1 values, indicating a prominent consequence resulted from β2AR-KO.
Numeric statistics of differentially expressed genes (DEGs) related with β2AR-KO in transgenic animal model of AD
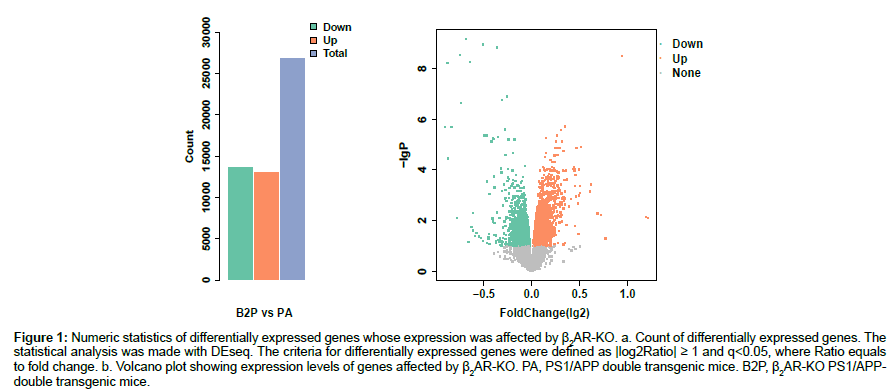
Raw data of the expression levels of the 45,281 transcripts were normalized to systematic differences between different BeadChips. Differential expression analysis on the normalized data was made by DEseq. Those genes whose normalized expression levels have absolute values of log2 ratio that are not less than one and a false discovery rate less than 0.05 (q<0.05) were taken as DEGs. Knock-out of β2AR gene significantly changed the expression levels of more than 50% of total genes manifested as the total transcripts probed, with down-regulated transcripts being more than up-regulated transcripts (Figures 1a, 1b).
Figure 1: Numeric statistics of differentially expressed genes whose expression was affected by β2AR-KO. a. Count of differentially expressed genes. The statistical analysis was made with DEseq. The criteria for differentially expressed genes were defined as |log2Ratio| ≥ 1 and q<0.05, where Ratio equals to fold change. b. Volcano plot showing expression levels of genes affected by β2AR-KO. PA, PS1/APP double transgenic mice. B2P, β2AR-KO PS1/APPdouble transgenic mice.
Hierarchical cluster analysis of DEGs related with β2AR-KO in transgenic animal model of AD
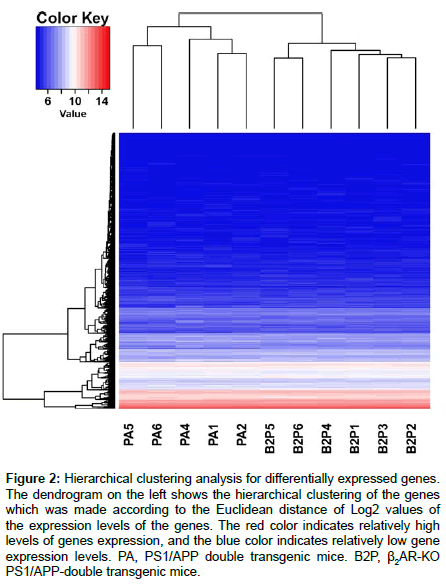
Hierarchical cluster analysis is a statistical approach to assign normalized DEG values into different clusters, with intra-cluster difference being much smaller than intercluster difference. It is different from classification in that it is an unsupervised mathematical grouping regardless of biological functions related with a normalized value. Phenotype structures of the samples are revealed by a hierarchical clustering figure. The R software was used by us to show two-dimensional hierarchical clustering. One dimension of the construction is the biological samples listed individually in horizontal, the other is DEGs identified in the above statistics. A dendrogram along the vertical axis showing hierarchical clustering of DEGs was achieved according to the Euclidean metrics of the Log2 values of the expression levels of DEGs (Figure 2). Zooming in the hierarchical clustering diagram, the most intuitive changes are that the DEGs with moderate transcriptional levels in PS1/APP mice are obviously decreased by β2AR-KO, and some other genes with relatively lower transcriptional levels are obviously increased.
Figure 2: Hierarchical clustering analysis for differentially expressed genes. The dendrogram on the left shows the hierarchical clustering of the genes which was made according to the Euclidean distance of Log2 values of the expression levels of the genes. The red color indicates relatively high levels of genes expression, and the blue color indicates relatively low gene expression levels. PA, PS1/APP double transgenic mice. B2P, β2AR-KO PS1/APP-double transgenic mice.
Gene ontology (GO) classification of DEGs related with β2AR-KO in transgenic animal model of AD
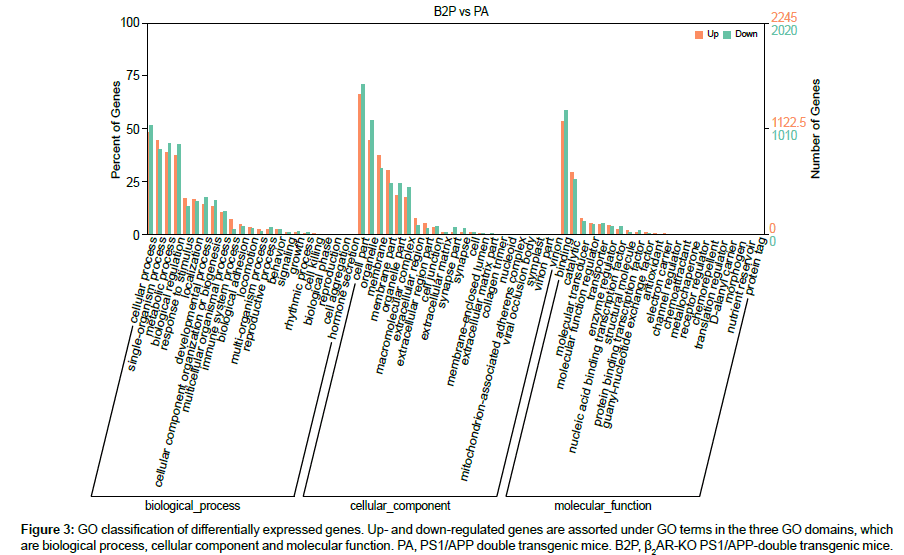
Currently, GO construction is aimed to pave the way to interpret or predict functions of genes or proteins by extracting and analyzing gene-related knowledges accumulated in large amount of literatures, leading to artificial intelligence. In GO consortium, there are three independent ontology domains, which are biological processes, cellular component and molecular function, using controlled vocabularies which are updated dynamically. In each of the three domains, there are subdomains to form tree structures with levels, nodes and relationships. According to the biological terms at the third level specified in the GO database typically, the expression of genes up- or down-regulated by β2AR-KO are classified and counted (Figure 3). There are four biological processes that are prominently affected by β2AR-KO, which are cellular process, single-organism process, metabolic process and biological regulation, each comprises more than 35% of total DEGs. In the domain of cellular components, gene expression levels that are mostly changed by β2AR-KO are cell part, membrane, membrane part, organelle, organelle part and macromolecular complex. In the domain of molecular function, the most prominent result is that the genes whose biological function is relavent to binding comprises more than 50% of total DEGs. At this step, these results primarily associated β2AR with membrane and its main function which is binding, which are indispensable for protein processing and presentation, which are immunological functions and shown in the following section.
Q-value distribution of GO terms enriched with DEGs related with β2AR-KO in transgenic animal model of AD
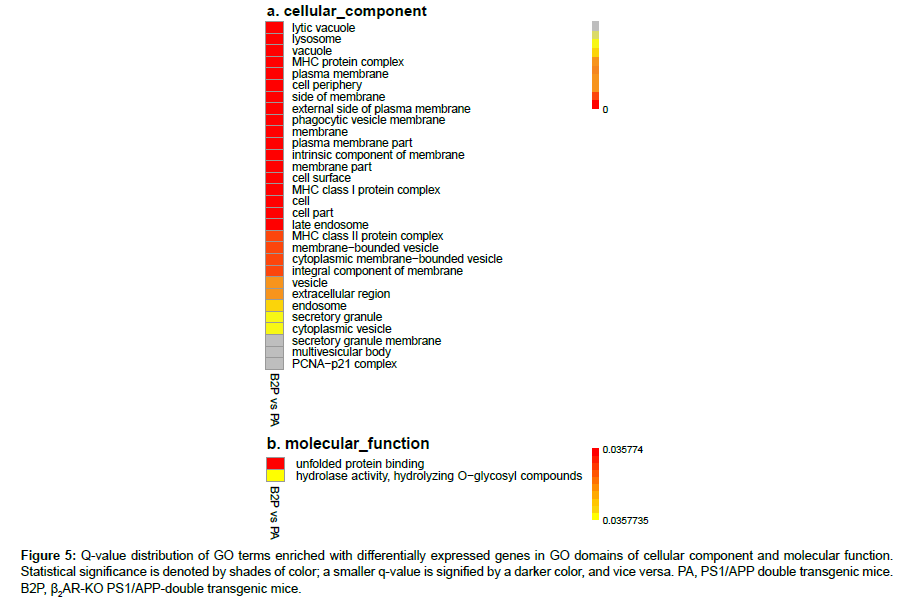
In order to control increased probability of type I error within all rejections associated with multiple hypothesis test of differential expression levels, false discovery rate (FDR) was calculated. The q-value is the minimum positive FDR (pFDR) to reject a null hypothesis for a given rejection region Γα of a GO term enriched with genes and an observed statistic T=t, and is defined as inf{Γα:t∈Γα} pFDR(Γα) (19). The distribution charts for GO term-enriched DEGs related with β2AR-KO were drawn according to q-values (Figures 4, 5a, 5b). The biological process (BP) domain comprises terms describing series of events accomplished by organized assemblies of molecular functions in living units. In GOBP, most of the terms enriched with DEGs are directly related with immunological function; there are 88 in total 117 terms are related, and 54 in 68 terms have very high strength which were highlighted in red (Figure 4). The top 3 highest enriched GO terms showed that β2AR primarily affects protein processing and presentation in the transgenic animal model of AD, which is pertinent to abnormal Aβ accumulation in the brain (Figure 4). Besides the top 3, twelve terms showed facets of the protein processing and presentation affected by β2AR-KO, including MHC class I and II mediating presentation of endogenous and exogenous proteins, peptides and even polysaccharides (Figure 4). In addition to the primary effects of β2AR on gene expression, other genes are also affected. There are six enriched GO terms related with protein folding, four related with nitric oxide synthesis, three related with response to stimulus, and ten related with chemical metabolism (Figure 4). Another two terms drawn attention are mRNA splice site selection and sphingolipid biosynthesis (Figure 4). In the GO domain of cellular component (CC), almost all of the terms are membranerelated events (27 out of total 30 terms), including MHC protein complex, plasma membrane, external side of plasma membrane, intrinsic component of membrane, endosome, vesicle, secretory granule and etc. (Figure 5a). The most prominent GOCC results are consistent with GOBP (Figure 5a). In the GO domain of molecular function, there are only two significantly enriched terms, and the most affected one is unfolded protein binding, which is a membrane function related with protein processing and presentation (Figure 5b).
Figure 4: Q-value distribution of GO terms enriched with differentially expressed genes in GO domain of biological process. Statistical significance is denoted by shades of color; a smaller q-value is signified by a darker color, and vice versa. PA, PS1/APP double transgenic mice. B2P, β2AR-KO PS1/ APP-double transgenic mice.
Figure 5: Q-value distribution of GO terms enriched with differentially expressed genes in GO domains of cellular component and molecular function. Statistical significance is denoted by shades of color; a smaller q-value is signified by a darker color, and vice versa. PA, PS1/APP double transgenic mice. B2P, β2AR-KO PS1/APP-double transgenic mice.
Hierarchical relationships of GO terms highly enriched with DEGs related with β2AR-KO in transgenic animal model of AD
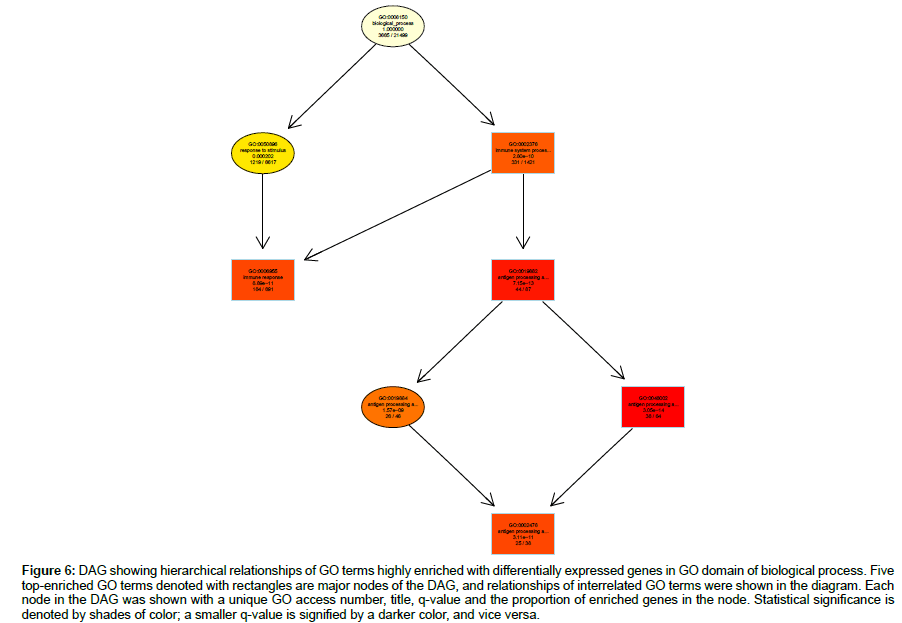
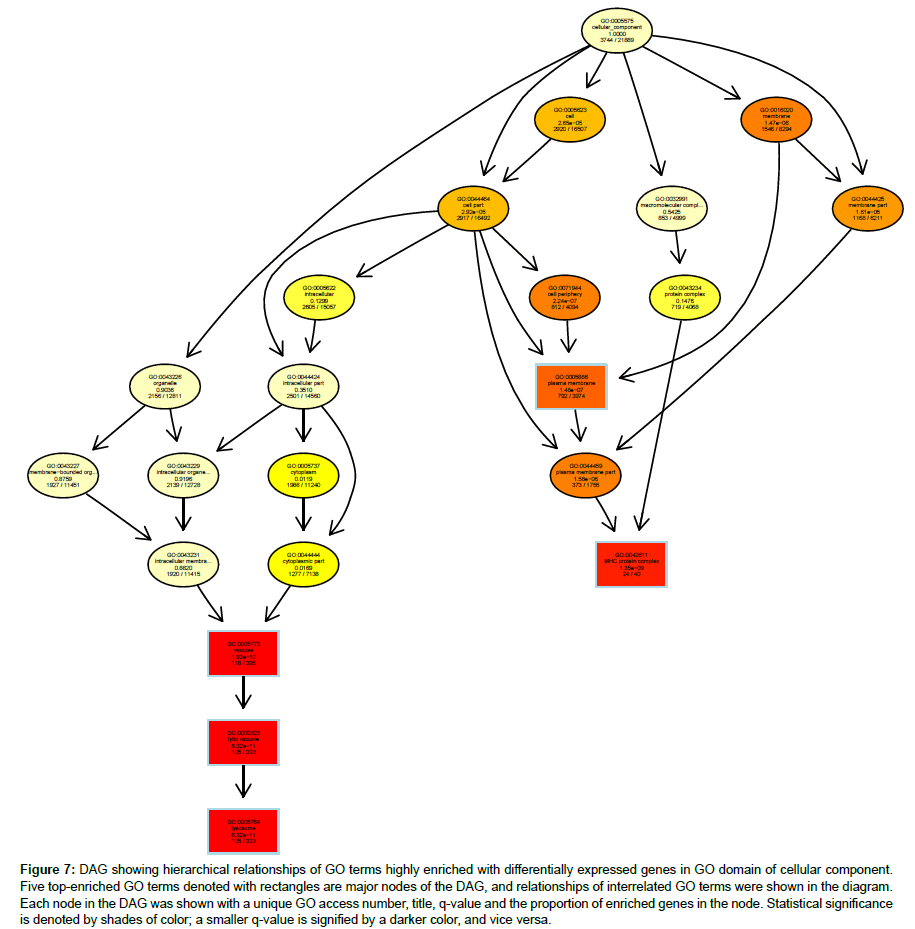
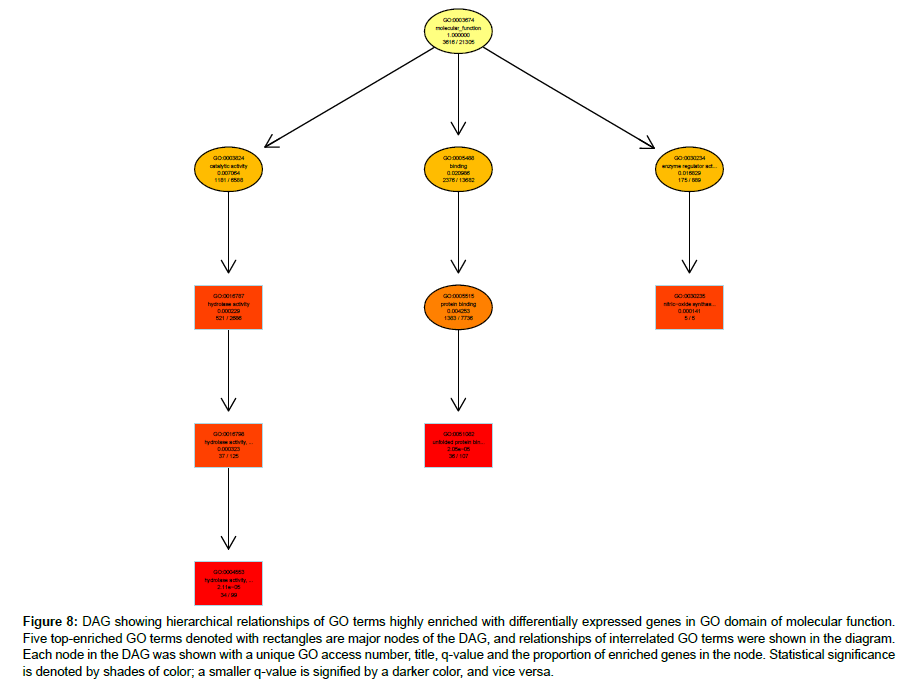
Hierarchical relationships among GO terms highly enriched with DEGs were shown by directed acyclic graphs (DAG) in detail (Figures 6-8). DAG was drawn based on automated analysis in each of the three independent GO ontologies which are biological process, cellular component and molecular function. In each of the three GO ontologies, top five DEGs-enriched GO terms are major nodes in DAG. In GOBP, all six highly enriched nodes are related to immune, with q-values ranging from 3.05 e-14 to 1.57 e-9, and 4 out of the 6 nodes showed that β2AR-KO fundamentally changed expression levels of genes regulating antigen processing (Figure 6). Top genes annotated in GO to be related to antigen processing and presentation were listed in the sequence of the fold of change (Table 1). In GOCC, the GO terms enriched with DEGs whose expression was affected by β2AR-KO are directed toward two directions. One group of GOCC nodes are directed toward MHC protein complex which was highly enriched with a q value of 1.3 e-9 through 3 paths, two of which are significantly enriched and related with membrane (Figure 7). The other group of GOCC nodes are directed toward a one-way path consisted of three consecutive nodes starting from vacuole (q=1.93 e-10), which in animals are typically founded in the protein-processing steps of endocytosis and exocytosis, and ending with lysosome (q=6.32 e-11) which is a component of protein processing and presentation (Figure 7). Top genes annotated in GO to be related to lysosome (Tables 2a and 2b) and MHC protein complex (Tables 3a and 3b) were listed in the sequence of the fold of change. In GOMF, there are 3 unrelated paths. One is the hydrolase activity path, which can be found in lysosomes and related to protein processing and presentation. The hierarchical node sequence in the hydrolase activity path is catalytic activity (GO:0003824, q=7.064 × 10-3, enrichment rate (ER):1181/6588), hydrolase activity (GO:0016787, q=2.29 × 10-4, ER: 521/2686), hydrolase activity acting on glycosyl bonds (GO:0016798, q=3.23 × 10-4, ER: 37/125) and hydrolase activity hydrolyzing O-glycosyl compounds (GO: 0004553, q=2.11 e-5, ER: 34/99) (Figure 8). Top genes annotated in GO to be related to hydrolase activity were listed in the sequence of the fold of change (Tables 4a and 4b). Another path is directed toward unfolded protein binding, which is a step of protein processing and presentation and may be related with AD pathology. The node sequence of the binding path is binding (GO: 0005488, q=2.0986 × 10-2, ER: 2376/13682), protein binding (GO: 0005515, q=4.253 × 10-3, ER: 1383/7736) and unfolded protein binding (GO: 0051082, q=2.05 e-05, ER: 36/107) (Figure 8). Top genes annotated in GO to be related to unfolded protein binding were listed in the sequence of the fold of change (Table 5). The third path is from the node of enzyme regulatory activity (GO: 0030234, q=1.6829 × 10-2, ER: 175/889), which is reserved for cases when the regulator directly interacts with the enzyme, to nitric-oxide synthase regulator activity (GO: 0030235, q=1.41 × 10-4, ER: 5/5) (Figure 8). Besides, by further reviewing of DEG data, it was found that the expression of all 5 genes comprised in GO nodes of nitric-oxide synthase regulator activity was decreased by β2AR-KO in the transgenic AD animal model. In addition to the above GO analysis, the genes that are annotated to be related to AD in the KEGG’s pathways were listed in Table 6, including Apolipoprotein E, which is the strongest genetic risk factor for both early- and late-onset AD found in one third of the cases of AD.
| Items | Gene ID | Fold Chg. | pvalue | qvalue |
|---|---|---|---|---|
| Histocompatibility 2, class II antigen A, beta 1 | 14961 | 8.871354 | 1.91E-06 | 0.000375 |
| CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | 16149 | 6.778798 | 2.57E-05 | 0.002088 |
| Cathepsin E | 13034 | 5.167045 | 2.08E-07 | 9.51E-05 |
| Histocompatibility 2, D region locus 1 | 14964 | 3.86412 | 6.03E-07 | 0.000192 |
| Histocompatibility 2, K1, K region | 14972 | 3.259361 | 3.27E-05 | 0.002469 |
| Proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 16912 | 2.908455 | 3.20E-05 | 0.00244 |
| Histocompatibility 2, class II, locus Mb1 | 14999 | 2.430133 | 5.14E-07 | 0.000173 |
| Histocompatibility 2, class II, locus Mb2 | 15000 | 2.132515 | 3.91E-08 | 2.70E-05 |
| Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 18173 | 2.006802 | 1.74E-05 | 0.001625 |
| Histocompatibility 2, O region alpha locus | 15001 | 1.993441 | 0.000614 | 0.01491 |
| Proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 16913 | 1.861686 | 0.000539 | 0.013966 |
| Fc receptor, IgE, high affinity I, gamma polypeptide | 14127 | 1.769161 | 8.53E-05 | 0.004534 |
| Beta-2 microglobulin | 12010 | 1.684328 | 4.52E-05 | 0.003045 |
| Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 21355 | 1.628326 | 0.003592 | 0.042868 |
| Fc receptor, IgG, low affinity Iib | 14130 | 1.62805 | 0.000151 | 0.006517 |
| Histocompatibility 2, M region locus 3 | 14991 | 1.626856 | 1.77E-05 | 0.001647 |
| Unc-93 homolog B1 (C. elegans) | 54445 | 1.538428 | 0.000216 | 0.00799 |
| Nucleotide-binding oligomerization domain containing 1 | 107607 | 1.503754 | 0.000728 | 0.016624 |
| Fc receptor, IgG, low affinity III | 14131 | 1.411944 | 0.001029 | 0.021051 |
| Fc receptor, IgG, alpha chain transporter | 14132 | 1.33175 | 0.003783 | 0.044161 |
Table 1: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to antigen processing and presentation in Gene Ontology.
| Items | Gene ID | Fold Chg. | pvalue | qvalue |
|---|---|---|---|---|
| CD74 antigen (invariant polypeptide of major | 16149 | 6.778798 | 2.57E-05 | 0.002088 |
| histocompatibility complex, class II antigen-associated) | ||||
| Cathepsin C | 13032 | 5.514072 | 0.002342 | 0.033643 |
| Interleukin 4 induced 1 | 100328588 | 2.760841 | 0.000134 | 0.006 |
| Solute carrier family 15, member 3 | 65221 | 2.480609 | 1.68E-07 | 8.88E-05 |
| CD68 antigen | 12514 | 2.203099 | 6.29E-06 | 0.000837 |
| Heparanase | 15442 | 2.188486 | 6.48E-07 | 0.000198 |
| RIKEN cDNA 5430435G22 gene | 226421 | 2.148704 | 4.72E-05 | 0.003128 |
| Histocompatibility 2, class II, locus Mb2 | 15000 | 2.132515 | 3.91E-08 | 2.70E-05 |
| Alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB) | 27419 | 2.13038 | 6.49E-06 | 0.000851 |
| Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 18173 | 2.006802 | 1.74E-05 | 0.001625 |
| Prolylcarboxypeptidase (angiotensinase C) | 72461 | 1.972664 | 5.00E-05 | 0.003226 |
| N-sulfoglucosamine sulfohydrolase (sulfamidase) | 27029 | 1.965037 | 1.81E-05 | 0.001656 |
| Glucuronidase, beta | 110006 | 1.954243 | 1.93E-05 | 0.001715 |
| Cathepsin Z | 64138 | 1.944783 | 4.78E-05 | 0.003156 |
| Lysosomal-associated protein transmembrane 5 | 16792 | 1.943501 | 2.60E-06 | 0.000457 |
| Mannosidase 2, alpha B1 | 17159 | 1.755669 | 6.14E-05 | 0.003691 |
| Arylsulfatase G | 74008 | 1.669844 | 0.00139 | 0.024861 |
| LPS-induced TN factor | 56722 | 1.644251 | 1.01E-05 | 0.001124 |
| Lysosomal-associated membrane protein 2 | 16784 | 1.628241 | 2.08E-06 | 0.000394 |
| Niemann Pick type C1 | 18145 | 1.623278 | 6.90E-06 | 0.000884 |
| CD63 antigen | 12512 | 1.619914 | 0.000166 | 0.006884 |
| Ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) | 12752 | 1.617276 | 5.01E-05 | 0.003226 |
| Galactosidase, beta 1 | 12091 | 1.616097 | 3.79E-05 | 0.00271 |
| Niemann Pick type C2 | 67963 | 1.614508 | 7.80E-06 | 0.00094 |
| Deoxyribonuclease II alpha | 13423 | 1.595048 | 0.00179 | 0.029031 |
| Unc-93 homolog B1 (C. elegans) | 54445 | 1.538428 | 0.000216 | 0.00799 |
Table 2a: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related lysozyme in Gene Ontology (Part I).
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| Cathepsin K | 13038 | 7.580496 | 0.000158 | 0.006652 |
| Cathepsin S | 13040 | 10.41212 | 0.001679 | 0.027926 |
| Sialidase 4 | 241159 | 5.623464 | 0.002891 | 0.037488 |
| Peroxiredoxin 6 | 11758 | 5.989792 | 0.002557 | 0.035172 |
| Syntaxin 8 | 55943 | 7.56421 | 0.001079 | 0.021551 |
| Neuraminidase 1 | 18010 | 10.8563 | 0.000602 | 0.014729 |
| Solute carrier family 29 (nucleoside transporters), member 3 | 71279 | 8.107022 | 0.003121 | 0.039352 |
| RIKEN cDNA 0610031J06 gene | 56700 | 9.661221 | 0.000288 | 0.009493 |
| Iduronidase, alpha-L- | 15932 | 7.167215 | 0.003211 | 0.039979 |
| Immunity-related GTPase family M member 1 | 15944 | 7.71878 | 0.003203 | 0.039973 |
| ATP-binding cassette, sub-family B (MDR/TAP), member 9 | 56325 | 9.114246 | 0.001726 | 0.028518 |
| Corticotropin releasing hormone binding protein | 12919 | 11.26217 | 0.000188 | 0.007354 |
| Glucosidase, alpha, acid | 14387 | 11.3279 | 0.00054 | 0.013971 |
| Glucosidase, beta, acid | 14466 | 9.797483 | 0.002908 | 0.037603 |
| Phospholipase A2, group XV | 192654 | 10.78939 | 0.002593 | 0.035401 |
| Mannosidase, beta A, lysosomal | 110173 | 8.529202 | 0.000965 | 0.020127 |
| Ceroid-lipofuscinosis, neuronal 5 | 211286 | 9.777018 | 0.001512 | 0.026133 |
| Vacuolar protein sorting 11 (yeast) | 71732 | 9.435465 | 0.000712 | 0.016323 |
| Lysosomal-associated membrane protein 1 | 16783 | 12.1733 | 0.001168 | 0.022445 |
| Arylsulfatase B | 11881 | 11.80603 | 0.002461 | 0.034612 |
| VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) | 67048 | 10.57272 | 0.003456 | 0.041699 |
| Transmembrane protein 55A | 72519 | 11.26718 | 0.002119 | 0.031692 |
| Sphingosine kinase 2 | 56632 | 4.96189 | 0.000586 | 0.014558 |
| Iduronate 2-sulfatase | 15931 | 6.259901 | 0.00208 | 0.031336 |
| Transmembrane protein 74 | 239408 | 5.943605 | 2.08E-06 | 0.000394 |
Table 2b: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to lysozyme in Gene Ontology (Part II)
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| Histocompatibility 2, class II antigen A, beta 1 | 14961 | 8.871354 | 1.91E-06 | 0.000375 |
| CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated) | 16149 | 6.436063 | 9.77E-06 | 0.001104 |
| cathepsin E | 13034 | 5.167045 | 2.08E-07 | 9.51E-05 |
| Histocompatibility 2, D region locus 1 | 14964 | 3.86412 | 6.03E-07 | 0.000192 |
| Histocompatibility 2, Q region locus 8 | 15019 | 3.623889 | 1.40E-05 | 0.001408 |
| Histocompatibility 2, K1, K region | 14972 | 3.259361 | 3.27E-05 | 0.002469 |
| Predicted gene 8909 | 667977 | 3.114538 | 8.43E-07 | 0.000229 |
| Proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 16912 | 2.908455 | 3.20E-05 | 0.00244 |
| Histocompatibility 2, class II, locus Mb1 | 14999 | 2.430133 | 5.14E-07 | 0.000173 |
| Eukaryotic translation initiation factor 2-alpha kinase 2 | 19106 | 2.220892 | 4.71E-07 | 0.00016 |
| Histocompatibility 2, class II, locus Mb2 | 15000 | 2.132515 | 3.91E-08 | 2.70E-05 |
| Lymphocyte-activation gene 3 | 16768 | 2.047944 | 0.000108 | 0.005212 |
| Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | 18173 | 2.006802 | 1.74E-05 | 0.001625 |
| Histocompatibility 2, O region alpha locus | 15001 | 1.993441 | 0.000614 | 0.01491 |
| Proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) | 16913 | 1.861686 | 0.000539 | 0.013966 |
| Fc receptor, IgE, high affinity I, gamma polypeptide | 14127 | 1.769161 | 8.53E-05 | 0.004534 |
| Histocompatibility 2, K1, K region | 14972 | 1.763206 | 2.74E-06 | 0.000472 |
| DnaJ (Hsp40) homolog, subfamily C, member 16 | 214063 | 1.713747 | 1.49E-05 | 0.001453 |
| beta-2 microglobulin | 12010 | 1.684328 | 4.52E-05 | 0.003045 |
| Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) | 21355 | 1.628326 | 0.003592 | 0.042868 |
| Fc receptor, IgG, low affinity Iib | 14130 | 1.62805 | 0.000151 | 0.006517 |
| Histocompatibility 2, M region locus 3 | 14991 | 1.626856 | 1.77E-05 | 0.001647 |
| Protein tyrosine phosphatase, non-receptor type 6 | 15170 | 1.62081 | 0.000859 | 0.018684 |
| Unc-93 homolog B1 (C. elegans) | 54445 | 1.538428 | 0.000216 | 0.00799 |
| Nucleotide-binding oligomerization domain containing 1 | 107607 | 1.503754 | 0.000728 | 0.016624 |
Table 3a: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to MHC protein complex in Gene Ontology (Part I).
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| Cyclin D1 | 12443 | 1.489153 | 0.000724 | 0.016556 |
| DnaJ (Hsp40) homolog, subfamily C, member 18 | 76594 | 1.461304 | 0.000678 | 0.015844 |
| CD3 antigen, epsilon polypeptide | 12501 | 1.42577 | 0.003388 | 0.04123 |
| Fc receptor, IgG, low affinity III | 14131 | 1.411944 | 0.001029 | 0.021051 |
| Fc receptor, IgG, alpha chain transporter | 14132 | 1.33175 | 0.003783 | 0.044161 |
| ATP-binding cassette, sub-family B (MDR/TAP), member 9 | 56325 | 1.305657 | 0.001726 | 0.028518 |
| Wolfram syndrome 1 homolog (human) | 22393 | 1.289007 | 0.003984 | 0.045336 |
| SEC63-like (S. cerevisiae) | 140740 | 1.193609 | 0.001819 | 0.029331 |
| HERPUD family member 2 | 80517 | -1.22882 | 0.003121 | 0.039352 |
| Heat shock protein 90, alpha (cytosolic), class A member 1 | 15519 | -1.23512 | 0.001109 | 0.021868 |
| Heat shock 105kDa/110kDa protein 1 | 15505 | -1.24793 | 0.001371 | 0.024635 |
| DnaJ (Hsp40) homolog, subfamily B, member 11 | 67838 | -1.29418 | 0.000546 | 0.014039 |
| Chaperonin containing Tcp1, subunit 3 (gamma) | 12462 | -1.35609 | 0.000138 | 0.006112 |
| Heat shock protein 90 alpha (cytosolic), class B member 1 | 15516 | -1.4133 | 0.000366 | 0.011053 |
| Heat shock protein 90, beta (Grp94), member 1 | 22027 | -1.41384 | 0.002119 | 0.031692 |
| T-complex protein 1 | 21454 | -1.41447 | 0.002478 | 0.034708 |
| Heat shock protein 5 | 14828 | -1.43609 | 0.001112 | 0.021872 |
| DnaJ (Hsp40) homolog, subfamily A, member 4 | 58233 | -1.46868 | 5.87E-05 | 0.00359 |
| DnaJ (Hsp40) homolog, subfamily C, member 3 | 1.00E+08 | -1.50583 | 0.000205 | 0.007744 |
| Serine/arginine-rich splicing factor 10 | 14105 | -1.54974 | 0.002451 | 0.034597 |
| Heat shock protein 1 (chaperonin) | 15510 | -1.76402 | 0.000238 | 0.008435 |
| Heat shock protein 8 | 15481 | -1.8216 | 0.000412 | 0.011816 |
| Calreticulin | 12317 | -1.86027 | 0.001086 | 0.021609 |
| p53 and DNA damage regulated 1 | 68559 | -15.3898 | 1.55E-12 | 5.96E-09 |
Table 3b: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to MHC protein complex in Gene Ontology (Part II).
| Items | Gene ID | Fold Chg. | pvalue | qvalue |
|---|---|---|---|---|
| Cathepsin C | 13032 | 5.514072 | 0.002342 | 0.033643 |
| Kallikrein related-peptidase 6 | 19144 | 5.377842 | 2.08E-07 | 9.51E-05 |
| Cathepsin E | 13034 | 5.167045 | 2.08E-07 | 9.51E-05 |
| Proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) | 16912 | 2.908455 | 3.20E-05 | 0.00244 |
| Plasminogen activator, urokinase | 18792 | 2.825956 | 1.57E-09 | 2.64E-06 |
| Lysozyme 1 | 17110 | 2.51725 | 1.58E-06 | 0.000345 |
| Chymotrypsin-like elastase family, member 1 | 109901 | 2.386791 | 1.55E-08 | 1.38E-05 |
| Ectonucleotide pyrophosphatase/phosphodiesterase 6 | 320981 | 2.239908 | 1.35E-07 | 7.72E-05 |
| CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 | 67655 | 2.216111 | 6.35E-06 | 0.000841 |
| Peptidyl arginine deiminase, type II | 18600 | 2.204623 | 9.15E-06 | 0.001061 |
| Heparanase | 15442 | 2.188486 | 6.48E-07 | 0.000198 |
| Dual specificity phosphatase 1 | 19252 | 2.12332 | 0.002496 | 0.034772 |
| Caspase 1 | 12362 | 2.095731 | 6.53E-05 | 0.00386 |
| Phosphatidic acid phosphatase type 2C | 50784 | 2.080684 | 1.81E-07 | 9.01E-05 |
| Ribonuclease, RNase A family 4 | 58809 | 2.073185 | 2.71E-07 | 0.000106 |
| Phospholipase D family, member 4 | 104759 | 2.053021 | 1.40E-06 | 0.000322 |
| Lysozyme 2 | 17105 | 2.031321 | 6.58E-06 | 0.000859 |
| Agmatine ureohydrolase (agmatinase) | 75986 | 2.030783 | 0.000198 | 0.007654 |
| ATP-binding cassette, sub-family B (MDR/TAP), member 1B | 18669 | 2.025545 | 3.16E-05 | 0.002415 |
| Cathepsin Z | 64138 | 2.001336 | 0.000111 | 0.0053 |
| Prolylcarboxypeptidase (angiotensinase C) | 72461 | 1.972664 | 5.00E-05 | 0.003226 |
| N-sulfoglucosamine sulfohydrolase (sulfamidase) | 27029 | 1.965037 | 1.81E-05 | 0.001656 |
| glucuronidase, beta | 110006 | 1.954243 | 1.93E-05 | 0.001715 |
| Suppression of tumorigenicity 14 (colon carcinoma) | 19143 | 1.915971 | 0.000214 | 0.007947 |
| Proprotein convertase subtilisin/kexin type 4 | 18551 | 1.914996 | 0.000861 | 0.0187 |
| 2',3'-cyclic nucleotide 3' phosphodiesterase | 12799 | 1.91379 | 8.14E-06 | 0.000968 |
| Proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) | 16913 | 1.861686 | 0.000539 | 0.013966 |
| paraoxonase 3 | 269823 | 1.86039 | 1.43E-05 | 0.001416 |
| Phospholipase C, gamma 2 | 234779 | 1.819061 | 9.23E-06 | 0.001066 |
| Mannosidase 2, alpha B1 | 17159 | 1.755669 | 6.14E-05 | 0.003691 |
Table 4a: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to hydrolase activity in Gene Ontology (Top 30 increase).
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 26900 | -2.10674 | 0.002199 | 0.032358 |
| Phosphodiesterase 4A, cAMP specific | 18577 | -1.98027 | 7.42E-05 | 0.004177 |
| Protein tyrosine phosphatase, receptor type, O | 19277 | -1.97666 | 0.00269 | 0.035996 |
| Proteasome (prosome, macropain) subunit, beta type 5 | 19173 | -1.96916 | 1.71E-05 | 0.001616 |
| Glucosamine-6-phosphate deaminase 1 | 26384 | -1.95682 | 4.02E-05 | 0.0028 |
| N-acyl phosphatidylethanolamine phospholipase D | 242864 | -1.93054 | 0.000285 | 0.009404 |
| acyl-CoA thioesterase 1 | 26897 | -1.86093 | 0.000536 | 0.013913 |
| Spastin | 50850 | -1.75118 | 9.36E-06 | 0.001069 |
| DIS3 mitotic control homolog (S. cerevisiae) | 72662 | -1.74871 | 0.000286 | 0.009435 |
| Ectonucleoside triphosphate diphosphohydrolase 6 | 12497 | -1.74754 | 5.98E-05 | 0.003631 |
| Rhomboid, veinlet-like 3 (Drosophila) | 246104 | -1.7434 | 2.35E-05 | 0.001954 |
| Acyl-CoA thioesterase 7 | 70025 | -1.73511 | 4.17E-05 | 0.002877 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | 13207 | -1.70454 | 0.000419 | 0.011946 |
| Spastin | 50850 | -1.7034 | 0.001148 | 0.022269 |
| Protease, serine, 53 | 330657 | -1.69693 | 0.001317 | 0.024066 |
| Esterase D/formylglutathione hydrolase | 13885 | -1.6703 | 1.03E-05 | 0.001143 |
| Mannan-binding lectin serine peptidase 2 | 17175 | -1.65176 | 0.00124 | 0.023313 |
| Haloacid dehalogenase-like hydrolase domain containing 2 | 76987 | -1.64692 | 9.52E-05 | 0.004837 |
| Bisphosphate 3'-nucleotidase 1 | 23827 | -1.64313 | 0.000246 | 0.008591 |
| OTU domain, ubiquitin aldehyde binding 2 | 68149 | -1.63286 | 0.000229 | 0.008222 |
| Endo/exonuclease (5'-3'), endonuclease G-like | 208194 | -1.58646 | 0.001819 | 0.029331 |
| Histocompatibility 13 | 14950 | -1.57331 | 0.000107 | 0.005193 |
| Iduronate 2-sulfatase | 15931 | -1.573 | 0.00208 | 0.031336 |
| YME1-like 1 (S. cerevisiae) | 27377 | -1.57208 | 0.000551 | 0.014096 |
| Adenosine deaminase-like | 75894 | -1.57155 | 0.002845 | 0.037159 |
| Angiogenin, ribonuclease, RNase A family, 5 | 11727 | -1.5621 | 0.000464 | 0.012644 |
| Inositol polyphosphate-4-phosphatase, type II | 234515 | -1.54742 | 0.004207 | 0.046872 |
| Expressed sequence AI464131 | 329828 | -1.54346 | 0.000783 | 0.017454 |
| Myotubularin related protein 2 | 77116 | -1.5103 | 0.003353 | 0.041002 |
| Alkaline ceramidase 2 | 230379 | -1.50696 | 8.64E-05 | 0.004576 |
Table 4b: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to hydrolase activity in Gene Ontology (Top 30 decrease).
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| Eukaryotic translation initiation factor 2-alpha kinase 2 | 19106 | 2.220892 | 4.71E-07 | 0.00016 |
| DnaJ (Hsp40) homolog, subfamily C, member 16 | 214063 | 1.713747 | 1.49E-05 | 0.001453 |
| Cyclin D1 | 12443 | 1.489153 | 0.000724 | 0.016556 |
| DnaJ (Hsp40) homolog, subfamily C, member 18 | 76594 | 1.461304 | 0.000678 | 0.015844 |
| Wolfram syndrome 1 homolog (human) | 22393 | 1.289007 | 0.003984 | 0.045336 |
| SEC63-like (S. cerevisiae) | 140740 | 1.193609 | 0.001819 | 0.029331 |
| HERPUD family member 2 | 80517 | -1.22882 | 0.003121 | 0.039352 |
| Heat shock protein 90, alpha (cytosolic), Class A member 1 | 15519 | -1.23512 | 0.001109 | 0.021868 |
| Heat shock 105kDa/110kDa protein 1 | 15505 | -1.24793 | 0.001371 | 0.024635 |
| DnaJ (Hsp40) homolog, subfamily B, member 11 | 67838 | -1.29418 | 0.000546 | 0.014039 |
| Chaperonin containing Tcp1, subunit 3 (gamma) | 12462 | -1.35609 | 0.000138 | 0.006112 |
| Heat shock protein 90 alpha (cytosolic), class B | 15516 | -1.4133 | 0.000366 | 0.011053 |
| Heat shock protein 90, beta (Grp94), member 1 | 22027 | -1.41384 | 0.002119 | 0.031692 |
| T-complex protein 1 | 21454 | -1.41447 | 0.002478 | 0.034708 |
| Heat shock protein 5 | 14828 | -1.43609 | 0.001112 | 0.021872 |
| DnaJ (Hsp40) homolog, subfamily A, member 4 | 58233 | -1.46868 | 5.87E-05 | 0.00359 |
| DnaJ (Hsp40) homolog, subfamily C, member 3 | 1.00E+08 | -1.50583 | 0.000205 | 0.007744 |
| Serine/arginine-rich splicing factor 10 | 14105 | -1.54974 | 0.002451 | 0.034597 |
| Heat shock protein 1 (chaperonin) | 15510 | -1.76402 | 0.000238 | 0.008435 |
| Heat shock protein 8 | 15481 | -1.8216 | 0.000412 | 0.011816 |
| Calreticulin | 12317 | -1.86027 | 0.001086 | 0.021609 |
| p53 and DNA damage regulated 1 | 68559 | -15.3898 | 1.55E-12 | 5.96E-09 |
Table 5: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to unfolded protein binding in Gene Ontology.
| Items | Gene ID | Fold Chg. | p value | q value |
|---|---|---|---|---|
| Cytochrome c oxidase, subunit VI a, polypeptide 2 | 12862 | 2.523729 | 1.44E-05 | 0.001417 |
| ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | 11937 | 1.512032 | 0.000439 | 0.012244 |
| Beta-site APP-cleaving enzyme 2 | 56175 | 1.506286 | 8.44E-05 | 0.004494 |
| Inositol 1,4,5-triphosphate receptor 2 | 16439 | 1.451202 | 0.000228 | 0.008213 |
| Lipoprotein lipase | 16956 | 1.413552 | 0.000202 | 0.007714 |
| Apolipoprotein E | 11816 | 1.35843 | 0.000376 | 0.01129 |
| Caspase 8 | 12370 | 1.336963 | 0.003373 | 0.041103 |
| Calpain 1 | 12333 | 1.311871 | 0.001259 | 0.023432 |
| NADH dehydrogenase (ubiquinone) flavoprotein 3 | 78330 | 1.231451 | 0.004652 | 0.049888 |
| ATP synthase, H+ transporting, mitochondrial F0 complex, subunit B1 | 11950 | -1.26575 | 0.004255 | 0.047107 |
| Guanine nucleotide binding protein, alpha q polypeptide | 14682 | -1.26842 | 0.003271 | 0.040466 |
| Phospholipase C, beta 4 | 18798 | -1.29642 | 0.000965 | 0.020124 |
| Anterior pharynx defective 1c homolog (C.elegans) | 68318 | -1.56577 | 0.000409 | 0.011797 |
| RIKEN cDNA 1500003O03 gene | 56398 | -1.58374 | 2.07E-05 | 0.001806 |
| Cytochrome c oxidase subunit VIIa polypeptide 2-like | 20463 | -10.3201 | 8.36E-14 | 1.12E-09 |
Table 6: Profiles of differentially expressed genes in β2AR-KO transgenic AD model annotated to be related to Alzheimer’s disease in KEGG’s pathways.
Figure 6: DAG showing hierarchical relationships of GO terms highly enriched with differentially expressed genes in GO domain of biological process. Five top-enriched GO terms denoted with rectangles are major nodes of the DAG, and relationships of interrelated GO terms were shown in the diagram. Each node in the DAG was shown with a unique GO access number, title, q-value and the proportion of enriched genes in the node. Statistical significance is denoted by shades of color; a smaller q-value is signified by a darker color, and vice versa.
Figure 7: DAG showing hierarchical relationships of GO terms highly enriched with differentially expressed genes in GO domain of cellular component. Five top-enriched GO terms denoted with rectangles are major nodes of the DAG, and relationships of interrelated GO terms were shown in the diagram. Each node in the DAG was shown with a unique GO access number, title, q-value and the proportion of enriched genes in the node. Statistical significance is denoted by shades of color; a smaller q-value is signified by a darker color, and vice versa.
Figure 8: DAG showing hierarchical relationships of GO terms highly enriched with differentially expressed genes in GO domain of molecular function. Five top-enriched GO terms denoted with rectangles are major nodes of the DAG, and relationships of interrelated GO terms were shown in the diagram. Each node in the DAG was shown with a unique GO access number, title, q-value and the proportion of enriched genes in the node. Statistical significance is denoted by shades of color; a smaller q-value is signified by a darker color, and vice versa.
Discussion
AD has baffled scientists ever since German psychiatrist Alois Alzheimer reported the first case of AD in 1906, the cause of which has been poorly understood until now [19,20], and there is neither treatment to intervene its development, nor particular measure to be effective in preventing or delaying the onset of AD [21].
Traditional amyloid hypothesis proposes that abnormal accumulation and aggregation of Aβ in the brain is the central event triggering neuronal degeneration in AD, which may be related to abnormal protein processing and presentation. Since the early days, the aggregated Aβ has been believed to be the causal factor of AD disrupting homeostasis of calcium ions in neurons and inducing apoptosis [22,23]. However, the driving force for abnormally increased accumulation and aggregation of Aβ in AD are still vague. In recent years, psychological stress has been identified to be a possible trigger for AD, and the roles of adrenergic receptors especially β2AR have attracted researchers’ notice. Partially because of the inconsistency in the localization of aggregated Aβ plaques and degenerated neurons, in recent years, the effects of soluble Aβ including Aβ dimers have been studied [5,17,24-26]. In recent studies on soluble Aβ, the role of β2AR in mediating some of the effects of soluble Aβ has been identified [5,6,17,26]. It has been found that polymorphism of β2AR gene may play a role in the pathogenesis of sporadic late - onset Alzheimer’s disease (LOAD) in that both the 16Gly allele and the 27Glu allele of the β2AR gene were associated with an increased risk of LOAD and there was a significant interaction with the apolipoprotein E gene ε4 allele, the presence of which markedly increases the incidence of LOAD (3). From a point of view, a study showed that activation of β2AR enhances γ-secretase activity and Aβ production, which requires agonist-induced internalization of both β2AR and PS1 in a complex to late endosomes and lysosomes, where Aβ production was increased [4]. Furthermore, cerebral amyloid plaques were increased by chronic treatment with β2AR agonists in animal model of AD [4]. A genetic study showed a complicated scenario: βAR may be related to AD through numerous factors, including human leukocyte antigen genes, the renin–angiotensin system, poly (adenosine diphosphateribose) polymerase 1, nerve growth factor, vascular endothelial growth factor, the reduced form of nicotinamide adenine dinucleotide phosphate, matrix metalloproteinases, mitogen-activated protein kinase pathways, prostaglandins, cyclooxygenase-2, and nitric oxide synthase [27]. In AD patients, there is the loss of neurons in the locus ceruleus (LC), however the density of β2AR is increased in the cortical laminae II, III, IV and V in AD patients [2], whereas the cortex is highly degenerated during the development of AD, and is the earliest region to have Aβ deposition [28]. The observed results indicated the association of β2AR with AD.
Major discoveries in the study first arose from the analysis of q-value distribution of GO terms enriched with DEGs that are related with β2AR-KO in the three GO domains, which are GOBP, GOCC and GOMF. The q-value introduced by John D. Storey in 2003 is a measure of the probability of type I error within all statistical rejections made for the test of differential expression of genes, instead of within all samples, and it is a multiple hypothesis testing quantity and a natural counterpart to the p-value [19,29]. The q-value is the minimum pFDR to reject the null hypothesis that β2AR-KO induces no change for a specified rejection region and an observed statistic [19,29]. FDR was calculated to control the probability of type I error which was increased when multiple hypothesis tests are applied to massive samples of DNA microarray. In the study, the q-values are indicated with each statistic when multiple hypothesis tests were made using FDR [19,29]. The q-value distribution analysis revealed major effects of β2AR-KO in the transgenic animal model of AD. In the first place, in GOBP most of the DEGs affected by β2AR-KO are related to immunological processes, in which the top 3 GO terms are antigen processing and presentation of peptide antigen, antigen processing and presentation, and antigen processing and presentation of endogenous peptide antigen. In GOCC, the notion was supported by that most of the terms are related to membrane structure and function, especially the MHC I and II protein complexes which are essential elements for endogenous and exogenous protein processing and presentation. In GOMF, the only two significantly enriched entries strengthened the above notion; one is unfolded protein binding, the other is hydrolase activity hydrolyzing O-glycosyl compounds. The GO synonym of binding is involved in both the posttranslational folding process aided by chaperones to form correct three-dimensional conformation and tagging of proteins for degradation. Some of misfolded proteins are degraded in cytosol. Aberrant folding processes and accumulation of misfolded proteins may be related with neurodegeneration including AD. The q-value distribution analysis in the three GO domains was the first step to show consistently that the immunological effects of β2AR-KO on protein processing and presentation for both endogenous and exogenous proteins.
DAGs showed in detail the hierarchical relationships of GO terms that were enriched with DEGs affected by β2AR-KO in the transgenic animal model of AD. In the three GO domains, which are GOBP, GOCC and GOMF, the hierarchical relationships were shown by networks connecting GO nodes, in each of which the genes are assorted using an updated and controlled GO vocabulary. In the domains, the effects of β2AR-KO on the expression of genes for protein processing and presentation was further interpretated showing the relationship between the genomic effects of β2AR-KO on membrane structures, hydrolysis of proteins in lysosome, presentation of processed protein debris through MHC protein complex. In GOBP, β2AR-KO affected not only immune response but also antigen processing and presentation. On one hand, the immune response was affected through both the response to stimulus and immune system process involved in the development and functioning of the immune system. The two aspects have been thought to be factors influencing the onset of AD. On the other hand, the four interconnected GO nodes showed that fundamental effects of β2AR-KO on genomic expression were to change antigen processing and presentation including proteins, peptides and lipids. In GOCC, the genomic effects of β2AR-KO on lysosome and MHC protein complex were revealed, which are cellular components required for protein processing and presentation. In addition to exogenous antigens, intracellular peptides can be presented to TCRs as potential foreign antigens by complexing with MHCs. MHC molecule and the processed antigen bound to it interact with both CD4/CD8 coreceptors and variable Ig-like domain of TCR on cell surface to trigger activation of T lymphocytes [30]. Proteins or peptides are processed and presented by two classical pathways: cytosolic MHC class I and endocytic MHC class II pathways. In cytosolic or endogenous pathway, any nucleated cell presents cytosolic proteins by MHC class I molecules, including not only endogenous peptides derived from defective translation or protein turn over but also heterogenous proteins resulted from microorganism infection or cancerous proteins degraded by proteosome. Upon recognizing MHC I molecules, natural killer (NK) cells are inhibited, therefore NK cells recognize stressed cells to induce apoptosis faster when there is a reduction of MHC class I molecules on the surface of stressed cells. In endocylic or exogenous pathway, antigen-presenting cells, such as dendritic cells and macrophages phagocytize antigens into phagosomes. After they matured to lysosomes, antigens are cleaved and processed by acidic hydrolases, then bind to MHC class II molecules on cell surface. The cleaved peptides bound to MHC molecules on lysosomal membrane and exhibiting immunodominance are trafficked to and externalized on cell surface for presentation [31]. In addition to the major genomic effects of β2AR on protein processing and presentation, in GOMF, there are three subfields directed to unfolded protein binding, hydrolase activity hydrolyzing O-glycosyl compounds and nitricoxide synthase regulator activity. The synonyms of unfolded protein binding comprise chaperone activities, including fimbrium-specific, glycoprotein-specific, histone-specific, ribosomal and tubulinspecific activities, and binding unfolded endoplasmic reticulum proteins. The parent term for “unfolded protein binding” is protein binding, the synonyms of which are protein amino acid binding, protein degradation tagging activity, protein tagging activity, protein folding chaperone and alpha-2 macroglobulin receptor-associated protein activity.
Conclusion
The analyzation in the transgenic animal models of AD showed genomic effects of β2AR-KO on immunological processes, which are mainly protein-processing and presentation activities involving membrane structure and MHC class I and II protein complex. Besides, protein folding and degradation participated by chaperones and misfolded protein tagging are also affected by β2AR-KO.
Acknowledgments
We thank Branden Skarpiak and Elaine Wu for assistance in genotyping of laboratory mice. This work was supported by a Natural Science Foundation of Hainan Province 314058 (D.W.), a National Institute of Heart, Blood, and Lung grant HL082846 (Y.K.X.), and grants from National Natural Science Foundation of China 31460226 (D.W.) and 31760246 (D.W.). D.W. is a recipient of a NARSAD award and an Illinois Department of Public Health grant.
References
- Murphy MP, LeVine H (2010) Alzheimer's disease and the Amyloid-β Peptide. J Alzheimers Dis 19: 311-323
- Kalaria RN, Andorn AC, Tabaton M, Whitehouse PJ, Harik SI, et al. (1989) Adrenergic receptors in aging and Alzheimer's disease: Increased β2-Receptors in prefrontal cortex and hippocampus. J Neurochem 53: 1772-1781
- Yu JT, Tan L, Ou JR, Zhu JX, Liu K, et al. (2008) Polymorphisms at the beta2-adrenergic receptor gene influence Alzheimer's disease susceptibility. Brain Res 1210: 216-222.
- Ni Y, Zhao X, Bao G, Zou L, Teng L, et al. (2006) Activation of beta2-adrenergic receptor stimulates gamma-secretase activity and accelerates amyloid plaque formation. Nat Med 12: 1390-1396.
- Wang D, Fu Q, Zhou Y, Xu B, Shi Q, et al. (2013) β2 adrenergic receptor, protein kinase A (PKA) and c-Jun N-terminal kinase (JNK) signaling pathways mediate tau pathology in Alzheimer disease models. J Biol Chem 288: 10298-10307.
- Wisely EV, Xiang YK, Oddo S3 (2014) Genetic suppression of β2-adrenergic receptors ameliorates tau pathology in a mouse model of tauopathies. Hum Mol Genet 23: 4024-4034.
- Gelber RP, Ross GW, Petrovitch H, Masaki KH, Launer LJ, et al. (2013) Antihypertensive medication use and risk of cognitive impairment: The Honolulu-Asia Aging Study. Neurol 81: 888-895.
- White LR, Gelber RP, Launer LJ, Zarow C, Sonnen J, et al. (2013) Three-Dimensional study of neuromuscular junctions (NMJ) in heterozygous R98C knock-in CMT1B mouse model by overexpression Neuregulin 1 Type III. Neurol 80: S44.005.
- Gelber RP, Ross GW, Petrovitch H, Masaki KH, Launer LJ, et al. (2013) Antihypertensive medication use and risk of cognitive impairment: The Honolulu-Asia aging study. Neurol 81: 888-895.
- Dupre DJ, Herbert TE, Jockers R (2012) GPCR signalling complexes: Synthesis, assembly, trafficking and specificity. Springer Netherlands.
- Wang D, Yuen EY, Zhou Y, Yan Z, Xiang YK (2011) Amyloid beta peptide-(1-42) induces internalization and degradation of β2 adrenergic receptors in prefrontal cortical neurons. J Biol Chem 286: 31852-31863.
- Gloerich M, Bos JL (2010) Epac: Defining a new mechanism for cAMP action. Annu Rev Pharmacol Toxicol 50: 355-375.
- Clapham DE, Neer EJ (1997) G protein beta gamma subunits. Annu Rev Pharmacol Toxicol 37: 167-203.
- Wickman KD, Iniguez-Lluhl JA, Davenport PA, Taussig R, Krapivinsky GB, et al. (1994) Recombinant G-protein βγ-subunits activate the muscarinic-gated atrial potassium channel. Nature 368: 255-257
- Kim D, Lewis DL, Graziadei L, Neer EJ, Bar-Sagi D, et al. (1989) G-protein beta Gamma-subunits activate the cardiac muscarinic K+-channel via phospholipase A2. Nature 337: 557-560.
- Khan SM, Sleno R, Gora S, Zylbergold P, Laverdure JP, et al. (2013) The expanding roles of Gβγ subunits in G Protein-coupled receptor signaling and drug action. Pharmacol Rev 65: 545-577
- Wang D, Govindaiah G, Liu R, De Arcangelis V, Cox CL, et al. (2010) Binding of amyloid β peptide to β2 adrenergic receptor induces PKA-dependent AMPA receptor hyperactivity. FASEB J 24: 3511-3521
- Oakley H, Cole SL, Logan S, Maus E, Shao P, et al. (2006) Intraneuronal β-Amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: Potential factors in amyloid plaque formation. J Neurosci 26: 10129-10140
- Storey JD, Tibshirani R (2003) Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol 224: 149-157.
- Burns A, Iliffe S (2009) Alzheimer’s disease. BMJ 338: b158
- Kawas CH (2006) Medications and diet: Protective factors for AD? Alzheimer Dis Assoc Disord 20: S89-96
- Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, et al. (2011) Alzheimer's disease. Lancet 377: 1019-1031
- Yankner BA, Duffy LK, Kirschner DA (1990) Neurotrophic and neurotoxic effects of amyloid beta protein: Reversal by tachykinin neuropeptides. Science 250: 279-282
- Lesne SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, et al. (2013) Brain amyloid-β oligomers in ageing and Alzheimer’s disease. Brain 136: 1383-1398
- Mc Donald JM, Savva GM, Brayne C, Welzel AT, Forster G, et al. (2010) The presence of sodium dodecyl sulphate-stable Aβ dimers is strongly associated with Alzheimer-type dementia. Brain 133: 1328-1341
- Wang D, Xiang YK (2011) β-adrenergic receptor, amyloid β-peptide, and Alzheimer's disease. Curr Top Membr 67: 205-228.
- Luong K, Nguyen LT (2013) The role of beta-adrenergic receptor blockers in Alzheimer’s disease. Am J Alzheimers Dis Other Demen 28: 427-439
- Ross JA, McGonigle P1, Van Bockstaele EJ (2015) Locus Coeruleus, norepinephrine and Aβ peptides in Alzheimer's disease. Neurobiol Stress 2: 73-84.
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100: 9440-9445
- Münz C (2016) Autophagy beyond intracellular MHC Class II antigen presentation. Trends Immunol 37: 755-763.
- Aderem A, Underhill DM (1999) Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 17: 593-623.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi