Research Article, J Appl Bioinforma Comput Biol Vol: 12 Issue: 6
Structural Analysis of Noncovalent Interactions in CDK2 Inhibitor Complexes
Vijina Chakkyarath*
Department of Bioinformatics, Bharathiar University, Coimbatore, Tamil Nadu, India.
- *Corresponding Author:
- Vijina Chakkyarath
Department of Bioinformatics,
Bharathiar University,
Coimbatore,
Tamil Nadu,
India;
E-mail: vbioinfo2009@gmail.com
Received date: 20 June, 2023, Manuscript No. JABCB-23-103292;
Editor assigned date: 23 June, 2023, PreQC No. JABCB-23-103292 (PQ);
Reviewed date: 07 July, 2023, QC No. JABCB-23-103292;
Revised date: 21 December, 2023, Manuscript No. JABCB-23-103292 (R);
Published date: 28 December, 2023, DOI: 10.4172/2329-9533.1000291
Citation: Chakkyarath V (2023) Structural Analysis of Noncovalent Interactions in CDK2 Inhibitor Complexes. J Appl Bioinforma Comput Biol 12:6.
Abstract
A huge number of ATP site directed, small molecular inhibitors have been synthesized and tested for finding their biological activity against different types of kinases more than the past three decades. Cyclin dependent kinases are also one of the significant targets for drug discovery. Three dimensional structures of 6 CDK2 ATP complexes and 50 CDK2 inhibitor complexes have taken from PDB and IC50 values available for 23 of these complexes. As binding to ATP site and the biological activity may be dependent on the different noncovalent interactions such as hydrogen bonds, hydrophobic bonds and electrostatic, Vaanderwaals such as other interactions. In the present work we have analyzed these interactions in the CDK2-ATP complexes as well as CDK2 inhibitor complexes. Based on these interactions we have developed multiple regression models to account for the experimentally observed IC50 values. We have made extensive analysis of the amino acids ATP contacts amino acids inhibitor contacts. Also the extend of similarity between the various ligands has been quantified using 2D and 3D analysis methods.
Keywords: Noncovalent interactions; Vaanderwaals; Cyclin-dependent kinases; Ligand; CDK2 inhibitor
Introduction
Kinases are become one of the most important classes of drug target with around 30 different kinase are being developed and investigated for cancer treatment [1]. Cyclin-Dependent Kinases (CDKs) are protein kinases with a cyclin subunit and it is essential for enzymatic activity [2]. It is present in all type of eukaryotes, and is having crucial roles in signaling pathways to control normal human cell functions and active merely when linked with a regulatory partner. Eukaryotic cells contain at most nine CDKs, and those are, CDK1, 2, 3, and 4, are openly involved in regulation of cell cycle [3-5]. CDKs are dependable for regulating cell division cycle, helping to make sure that the genome is replicated once per cell cycle and it is required for timing and order of cell division [6-8]. CDK2 is a major constituent of the CDK complex, and it is responsible for the transition of G1/S phase and it is a monomer comprised of a polypeptide chain consisting of 298 amino acid residues with mainly α-helix elements as well as a β-sheet terminus [9].
The activation CDK involves two‐step process and that requires phosphorylation and cycline binding in the T loop [10,11]. Overactivity or insufficient activity of CDKs or is linked with several tumors, for this reason it became an important target in anticancer and antiviral drug discovery [12]. CDK2 inhibitors show exciting potential activity as tumor suppressors. The inhibitors of kinases interact with the backbone motif and are the part of binding site [8]. Finn, et al., in their article provides the most recent approaches of targeting this essential cell cycle regulatory mechanism in the perspective of breast cancer therapy [13].
Nonbonding interactions among proteins and ligands play essential roles in important biological processes mainly signal transduction and enzymatic reactions. Understanding these interactions is important for designing synthetic inhibitors. In this work, we focus on non-covalent interactions of 6 CDK2 ATP complexes and 50 CDK2 inhibitor complexes and discussed structural analysis of these interactions in 56 CDK complexes. Analyzing Structural information can be helpful for understanding these complexes at the molecular level.
Materials and Methods
The crystallographic data of 56 CDK complexes taken from the Protein Data bank (PDB) forms the source of present study. Amoung them 50 complexes belong to the small molecular inhibitors of CDK2 and 6 are ATP complexes [14]. The PDB ID for the proteins used in the present study along with the amino acid contacts are given in corresponding tables.
Analysis of protein ligand interactions
Each of the CDK2 complex structures were analyzed using the Java tool provided at the protein data bank website, which gives the details about different types of contacts such as bridged hydrogen bonds, hydrophobic bonds, hydrophilic bonds and other interactions [15]. The cutoff limits for the bridged hydrogen bonds is the distances between the ligand atoms and all H2O atoms in the structure and returns all the distances that are less than 5 A, for hydrophilic bonds the distances between potential H-bonds donors or accepters and returns all the distances that are within the range of set to 2.7 A for the limit and the upper limit is set to 3.3 A. For hydrophobic bonds the distances between C-C and returns all the distances that are within the range of lower limit is set to. 9 A, and the upper limit is set to 3.9 A, for other interactions the distances between the ligand atoms and protein atoms which are not between potential h-bonds donor or acceptors or C-C within the range of the lower limit is set to. 9 A and the upper limit is set to 3.9 A.
2D similarity
Here 2D comparison have done using the tool super ligands [16].This program will search for ligands similar to a given ligand and also compare two ligands by computing the Tanimoto coefficient. Here 23 small molecular inhibitors which have similar IC50 values have been taken for analysis.
3D superposition
In the similar way 3D superposition among the ligands has been done using the tool Super ligands. This program comparing all instances of two PDB ligands by performing a three dimensional and giving best fitting pair of ligands. The results section of this program shows number of atoms of structure 1, number atoms of structure 2, and number of superposed atoms, number of superposed atoms same type root mean square distance and superposed structure. Here root mean square values of 23 small molecular inhibitors of CDK have been chosen.
Single correlation and regression analysis
A correlation is a number between -1 and +1 that measures the degree of association between two variables. Positive value for the correlation implies a positive association and negative value implies a negative association or inverse association. In statistics regression analysis examines the relation of a dependent variable to specified independent variables. The mathematical model of their relationship is the regression equation. Uses of regression include curve fitting, modeling of casual relationships, and testing scientific hypothesis about relationship between variables. In the present work regression analysis were carried out for four processes.
• Interaction with binding energy values.
• Back check prediction value.
• Jack knife test.
• Amino acid ligand interactions.
Results and Discussions
Analysis of protein ligand interactions
A dataset of three dimensional structures of six CDK2 ATP complexes and 50 CKD2 inhibitor complexes are taken from PDB with reference to and IC50 values are available for 23 out of 50 CDK2 inhibitor complexes and are retrieved from Binding database [17,18]. The details of 6CDK2-ATP complexes and 50 CDK2 inhibitor complexes are listed in Table 1. 23 CDK2 inhibitor complexes out 50 that have IC50 values are listed in Table 2.
| PDBID | R | Sub-type | Cyclin | SMI | PI | ATP | References |
|---|---|---|---|---|---|---|---|
| 1b38 | 2 | CDK2 | - | - | - | Yes | Brown, et al. |
| 1b39 | 2.1 | CDK2 | - | - | - | Yes | Brown, et al. |
| 1fi n | 2.3 | CDK2 | CyclinA | - | - | Yes | Jeffrey, et al. |
| 1fq1 | 3 | CDK2 | - | - | - | Yes | Song, et al. |
| 1hck | 1.9 | CDK2 | - | - | - | Yes | Schulze-Gahmen, et al. |
| 1jst | 2.6 | CDK2 | CyclinA | - | - | Yes | Russo, et al. |
| 1AQ1 | 2 | CDK2 | - | Yes | - | - | Lawrie, et al. |
| 1CKP | 2.05 | CDK2 | - | Yes | - | - | Gray, et al. |
| 1D18 | 2.2 | CDK2 | - | Yes | - | - | Shewchuk, et al. |
| 1PXO | 1.96 | CDK2 | - | Yes | - | - | Wang, et al. |
| 1E1V | 1.95 | CDK2 | - | Yes | - | - | Arris, et al. |
| 1E1X | 1.85 | CDK2 | - | Yes | - | - | Arris, et al. |
| 1E9H | 2.5 | CDK2 | CyclinA3 | Yes | - | - | Davies, et al. |
| 1FVT | 2.2 | CDK2 | - | Yes | - | - | Davis, et al. |
| 1FVV | 2.8 | CDK2 | CylinA | Yes | - | - | Davis, et al. |
| 1G5S | 2.61 | CDK2 | - | Yes | - | - | Dreyer, et al. |
| 1GIH | 2.8 | CDK2 | - | Yes | - | - | Ikuta, et al. |
| 1GII | 2 | CDK2 | - | Yes | - | - | Ikuta, et al. |
| 1GIJ | 2.2 | CDK2 | - | Yes | - | - | Ikuta, et al. |
| 1GZ8 | 1.3 | CDK2 | - | Yes | - | - | Gibson, et al. |
| 1H0V | 1.9 | CDK2 | - | Yes | - | - | Gibson, et al. |
| 1H0W | 2. 10 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1H1P | 2. 10 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1H1Q | 2.5 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1H1R | 2 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1H1S | 2 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1JSV | 1.96 | CDK2 | - | Yes | - | - | Davies, et al. |
| 1H07 | 1.85 | CDK2 | - | Yes | - | - | Beattie, et al. |
| 1KE5 | 2.2 | CDK2 | - | Yes | - | - | Bramson, et al. |
| 1KE6 | 2 | CDK2 | - | Yes | - | - | Bramson, et al. |
| 1KE7 | 2 | CDK2 | - | Yes | - | - | Bramson, et al. |
| 1KE8 | 2 | CDK2 | - | Yes | - | - | Bramson, et al. |
| 1KE9 | 2 | CDK2 | - | Yes | - | - | Bramson, et al. |
| 1OGU | 2.6 | CDK2 | CyclinA | Yes | - | - | Sayle, et al. |
| 1OI9 | 2. 10 | CDK2 | CyclinA | Yes | - | - | Hardcastle, et al. |
| 1OIQ | 2.31 | CDK2 | - | Yes | - | - | Anderson, et al. |
| 1OIR | 1.91 | CDK2 | - | Yes | - | - | Anderson, et al. |
| 1OIT | 1.6 | CDK2 | CyclinA | Yes | - | - | Anderson, et al. |
| 1OIU | 2 | CDK2 | CyclinA | Yes | - | - | Hardcastle, et al. |
| 1OIY | 2.4 | CDK2 | CyclinA | Yes | - | - | Hardcastle, et al. |
| 1P2A | 2.5 | CDK2 | - | Yes | - | - | Liu, et al. |
| 1PF8 | 2.51 | CDK2 | - | Yes | - | - | Moshinsky, et al. |
| 1PKD | 2.3 | CDK2 | CyclinA | Yes | - | - | Johnson, et al. |
| 1PXI | 1.95 | CDK2 | - | Yes | - | - | Wu, et al. |
| 1PXJ | 2.3 | CDK2 | - | Yes | - | - | Wu, et al. |
| 1PXK | 2.8 | CDK2 | - | Yes | - | - | Wu, et al. |
| 1PXL | 2.5 | CDK2 | - | Yes | - | - | Wu, et al. |
| 1PXM | 2.53 | CDK2 | - | Yes | - | - | Wang, et al. |
| 1PXN | 2.5 | CDK2 | - | Yes | - | - | Wang, et al. |
| 1PXP | 2.3 | CDK2 | - | Yes | - | - | Wang, et al. |
| 1PYE | CDK2 | - | Yes | - | - | Hamdouchi, et al. | |
| 1R78 | 2 | CDK2 | - | Yes | - | - | Luk, et al. |
| 1URW | 2 | CDK2 | - | Yes | - | - | Byth, et al. |
| 1H08 | 1.8 | CDK2 | - | Yes | - | - | Beattie, et al. |
| 1HOO | 1.6 | CDK2 | - | Yes | - | - | Beattie, et al. |
| 1HO1 | 1.79 | CDK2 | - | Yes | - | - | Beattie, et al. |
Table 1: CDK2 inhibitor complexes.
| PDBID | IC50 | PDBID | IC50 |
|---|---|---|---|
| 1E1V | 17000 | 1KE6 | 5.7 |
| 1E1X | 2200 | 1KE7 | 8.9 |
| 1GII | 78 | 1KE8 | 1000 |
| 1GIJ | 25000 | 1KE9 | 660 |
| 1H00 | 38000 | 1OGU | 34 |
| 1H01 | 1000 | 1OIR | 32 |
| 1H1Q | 970 | 1OIT | 3 |
| 1H1R | 2300 | 1P2A | 12 |
| 1H1S | 5.4 | 1PYE | 324 |
| 1H07 | 3000 | 1R78 | 3 |
| 1H08 | 300 | 1URW | 3 |
| 1KE5 | 560 |
Table 2: List of 23 CDK2 inhibitor complexes with IC50 values.
2D similarity
Here 23 small molecular inhibitor which have IC50 value has been taken for analysis. Here we compared all 23 ligands with each other that have different activity. 23 small inhibitors with their hetro ID and 2D similarity values are shown in Table 3. Using this percentage values we can say that how much one ligand structure shows similarity with another ligand. Ligands that show best (maximum Tanimoto coefficients) are shown in bold [20]. The similarity and activity values of two ligand molecules are not always related. If the two molecules are of the same type, as the similarity increases the activity value may also show similar values. From the Table 3, for the same type of molecules 1H1Q and 1H1R have activity values 970 and 2300 respectively and they also show similarity of 97.76% but here it is also seen in the table that the different types of molecules such as 1H01 and 1KE8 having same activity value (1000, 1000) shows only 42% similarity [21].
| Amino acids | Total number | Number of occurrence | Percentage |
|---|---|---|---|
| I 10 | 6 | 6 | 100 |
| G 11 | 6 | 3 | 50 |
| E 12 | 6 | 4 | 67 |
| G 13 | 6 | 6 | 100 |
| T 14 | 6 | 4 | 67 |
| Y 15 | 6 | 3 | 50 |
| G 1 6 | 6 | 2 | 33 |
| V 18 | 6 | 5 | 83 |
| A3 1 | 6 | 5 | 83 |
| K33 | 6 | 5 | 83 |
| V64 | 6 | 5 | 83 |
| F80 | 6 | 3 | 50 |
| E8 1 | 6 | 6 | 100 |
| F82 | 6 | 6 | 100 |
| L83 | 6 | 6 | 100 |
| D86 | 6 | 4 | 67 |
| D 127 | 6 | 2 | 33 |
| F 146 | 6 | 1 | 17 |
| K89 | 6 | 1 | 17 |
| K 129 | 6 | 4 | 67 |
| G 147 | 6 | 1 | 17 |
| Q 13 1 | 6 | 4 | 67 |
| N 132 | 6 | 5 | 83 |
| L 134 | 6 | 5 | 83 |
| D 145 | 6 | 5 | 83 |
| A 149 | 6 | 1 | 17 |
Table 3: Representation of percentage of occurrence in various amino acid residues in ATP complexes.
3D superposition
In a similar manner 3D superposition also has been done for 23 CDK2 complexes which already have IC50 values. Here root mean square values of 23 small molecule inhibitors of CDK have been chosen. The activity is more in case of molecules having minimum root mean square value [22-30]. Ligands that shows best (minimum RMS deviations) are shown in bold for example in Table 4 the ligand 1H1Q and 1H1R having activity values 970 and 2300 shows minimum RMS deviation.
| Amino acids | Total number | Number of occurrence | Percentage |
|---|---|---|---|
| I 10 | 50 | 47 | 94 |
| G 11 | 50 | 15 | 30 |
| E 12 | 50 | 20 | 40 |
| G 13 | 50 | 12 | 24 |
| T 14 | 50 | 1 | 2 |
| V18 | 50 | 34 | 68 |
| A31 | 50 | 43 | 86 |
| K33 | 50 | 28 | 56 |
| V64 | 50 | 29 | 58 |
| F80 | 50 | 42 | 84 |
| E81 | 50 | 47 | 94 |
| F82 | 50 | 41 | 82 |
| L82 | 50 | 1 | 2 |
| H82 | 50 | 2 | 4 |
| L83 | 50 | 48 | 96 |
| V83 | 50 | 2 | 4 |
| H84 | 50 | 30 | 60 |
| T89 | 50 | 1 | 2 |
| Q85 | 50 | 21 | 42 |
| K89 | 50 | 19 | 38 |
| N132 | 50 | 14 | 28 |
| K88 | 50 | 1 | 2 |
| D86 | 50 | 35 | 70 |
| Q131 | 50 | 20 | 40 |
| L134 | 50 | 47 | 94 |
| A 144 | 50 | 18 | 36 |
| D145 | 50 | 39 | 78 |
| F146 | 50 | 10 | 20 |
| L298 | 50 | 1 | 2 |
Table 4: Representation of percentage of occurrence in various amino acid residues in small molecule inhibitors.
Comparison of contacts maps of two related molecules
Contacts map of two related complexes which have similar IC50 values are drawn here using the JAVA tool provided at the PDB site. Using contact maps we can find various amino acids residues that are in contacts with sugar molecules, bases adenine moieties and also extract various information about ligands and proteins interactions. Here we analyzed various nonlocal interactions present in the ligand [31]. Here two small molecule inhibitors of cyclin dependent kinase which have similar IC50 are compared. It is seen that various contacts are represented using various colors green for hydrophilic, pink for hydrophobic, blue for bridged hydrogen bond and white color for other contacts. In 1H1Q amino acid residues such as I10, E12, G13, V18, A31, V64, F80, E81, F82, 83, H84, Q85, K89, N82, 83, H84, Q85, K89, D86, Q82, 83, H84, Q85, K89, Q131, L134, A144, D145 are making contacts with small molecular inhibitors. In 1H1R amino acids residues such as I10, E12, A31, V64, F80, E81, F82, L83, H84, Q85, K89, N132, D86, Q131, L134 AND D145 are making contacts with small molecular inhibitor. In H1Q7 bridged hydrogen bonds, hydrophilic bonds, 22 hydrophobic bonds 28 other interactions are present. In 1H1R 22 bridged hydrogen bonds, 1 hydrophilic bond 34 hydrophobic bonds and 37 other interactions present. The comparison results shows that the relative molecules which have similar IC50 values have almost similar number of hydrophilic bonds, number of hydrophobic bonds and other nonlocal interactions but in case of bridged hydrogen bonds variation can be seen.
Contacts with the aminoacid residues
The ATP binding pocket: Aminoacid-ATP interaction: In Table 4 the amino acids that interact with the various atoms of ATP are iven. This table reveals that the residues I10, G13, E81, F82, and L83 are making contacts with ATP in all the ATP-CDK2 complexes.
Further in five out of six complexes the residues V18, A31, K33, V64, N132, L134 and D145 are also making interactions with ATP. Four additional contacts are observed from residues E12, T14, D86, K129 and Q131. The interacting residues further grouped on the basis of the interactions with the adenine, ribose and phosphate moieties of ATP. They are represented in this Table in various colors: Green color represents the sugars, blue for bases red for phosphate and brown for other contacts [32]. From the results we can that I10, A31, K33, V64, E81, F82, L82, L83, H84, D86, L134 are making contacts with the adenine moieties of ATP. Quite interestingly out of eleven residues seven are hydrophobic which imply that the adenine moiety is in hydrophobic environment. Residues G13, T14, K129, Q131, N132, and D145 are making contacts with the phosphate group [33].
The small molecule inhibitor complexes
In Table 5 amino acids that interact with small molecular inhibitors are given. The Table results reveals that the residues I10, A31, E81, F80, F82,L83 and l34 are making contacts with small molecular inhibitors. Further we can see that G11, E12, V18, V64, H84, K89, D86, F145 are making contacts with small molecular inhibitors in a partial manner. As described in Table 4 here also interacting residues are further classified on the basis of interactions with the adenine and ribose phosphate moieties. Color representation is similar to that the Table 4. In addition brown color represents new contacts. From the results we could observe that the small molecular inhibitors interact with the CDK2 in a manner similar to its natural ligand ATP. In addition new contacts with residues E8, K9, F80, Q85, N132, Q131, A144 and D145 are making contacts with some of the complexes. Differences and similarities in these interactions are expected to provide a rational for the varied IC50 values [34,35].
| Amino acids residues | Inhibitor | ATP |
|---|---|---|
| I10 | 94 | 100 |
| G11 | 30 | 50 |
| E12 | 40 | 67 |
| G13 | 50 | 100 |
| T14 | 2 | 67 |
| Y15 | 0 | 50 |
| G16 | 0 | 33 |
| V18 | 68 | 83 |
| A31 | 86 | 83 |
| K33 | 56 | 83 |
| V64 | 58 | 83 |
| F80 | 84 | 50 |
| E81 | 94 | 100 |
| F82 | 82 | 100 |
| L83 | 96 | 100 |
| H84 | 60 | 67 |
| D86 | 70 | 33 |
| K89 | 38 | 17 |
| D127 | 0 | 17 |
| Q131 | 40 | 67 |
| N132 | 28 | 17 |
| L134 | 94 | 67 |
| A144 | 36 | 83 |
| D145 | 78 | 83 |
| F146 | 20 | 83 |
Table 5: Combination of percentage of ATP complexes and small molecular inhibitors with various amino acid residues.
Representation of percentage of occurance in various aminoacid residues in ATP and SMI complexes
The percentage of occurrence of various amino acids in the binding pocket of ATP complexes are represented in Table 5. We observed that the residues such as I10, G13 and E81, F82, and L83 are having maximum number of percentage of occurrence.
The percentage of occurrence of various amino acids in the binding pocket of small molecule inhibitors is represented in Table 6. A similar correlation in the small molecule inhibitor shows that have maximum percentage of occurrence compare to other amino acid residues.
| S.no. | PDBID | Ligand name | No. of bridged hydrogen bonds | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total |
|---|---|---|---|---|---|---|---|
| 1 | 1B38 | ATP | 57 | 13 | 7 | 43 | 120 |
| 2 | 1B39 | ATP | 61 | 13 | 7 | 43 | 124 |
| 3 | 1FIN | ATP | 33 | 4 | 8 | 19 | 64 |
| 4 | 1FQ1 | ATP | ----- | 8 | 6 | 37 | 51 |
| 5 | 1HCK | ATP | 45 | 15 | 7 | 47 | 114 |
| 6 | 1JST | ATP | 9 | 9 | 10 | 56 | 84 |
Table 6: Ligand protein interactions in CDK2-ATP complexes.
Combination of percentage of ATP complexes and small molecular inhibitors with various aminoacid residues
Combination of percentage of ATP complexes and small molecular inhibitors with various amino acid contacts shown in Table 7. A graph displays the percentage of occurrence of amino acid in ATP complexes and small molecular inhibitor complexes.
| S.No | PDBID | No. of bridged hydrogen bonds | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total |
|---|---|---|---|---|---|---|
| 1 | 1AQ1 | 55 | 4 | 22 | 28 | 109 |
| 2 | 1CKP | 15 | 1 | 19 | 26 | 61 |
| 3 | 1D18 | 13 | 2 | 22 | 25 | 62 |
| 4 | 1DM2 | 20 | 5 | 10 | 22 | 57 |
| 5 | 1FVT | 8 | 4 | 19 | 33 | 64 |
| 6 | 1FVV | 9 | 4 | 26 | 39 | 78 |
| 7 | 1G5S | ----- | 4 | 22 | 17 | 43 |
| 8 | 1GIH | ----- | 1 | 9 | 22 | 32 |
| 9 | 1GII | 11 | 2 | 13 | 23 | 49 |
| 10 | 1GIJ | 28 | 2 | 9 | 29 | 68 |
| 11 | 1GZ8 | 43 | 3 | 13 | 23 | 82 |
| 12 | 1H00 | 28 | 1 | 18 | 32 | 79 |
| 13 | 1H0V | 31 | 4 | 8 | 16 | 59 |
| 14 | 1H0W | 3 | 1 | 13 | 13 | 30 |
| 15 | 1H1P | ----- | ------ | ----- | 25 | 25 |
| 16 | 1H1Q | 7 | 1 | 22 | 28 | 58 |
| 17 | 1H1R | 21 | 1 | 34 | 37 | 93 |
| 18 | 1H1S | 45 | 4 | 16 | 36 | 101 |
| 19 | 1JSV | 39 | 3 | 9 | 26 | 77 |
| 20 | 1H07 | 45 | 4 | 16 | 36 | 101 |
| 21 | 1H08 | 50 | 4 | 14 | 27 | 95 |
| 22 | 1KE5 | 27 | 2 | 16 | 24 | 69 |
| 23 | 1KE6 | 20 | 4 | 16 | 31 | 71 |
| 24 | 1KE7 | 12 | 5 | 10 | 36 | 63 |
| 25 | 1KE8 | 13 | 3 | 22 | 31 | 69 |
| 26 | 1KE9 | 15 | 2 | 14 | 10 | 41 |
| 27 | 1OGU | 21 | 1 | 16 | 32 | 70 |
| 28 | 1OI9 | 3 | 5 | 8 | ||
| 29 | 1OIR | 18 | 3 | 11 | 19 | 51 |
Table 7: Ligand protein interactions with CDK2-SMI complexes.
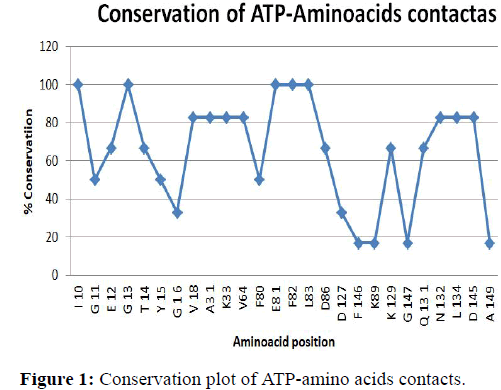
Conservation plot of amino acids contacts
Conservation plot of amino acid interactions CDK2-ATP complexes: In Figure 1 conservation plot of ATP-CDK2 complexes have shown. The amino acids that interact with ATP are taken in the X-axis and percentage of conservation is taken as Y-axis. Graph has drawn using Table 5 [36]. From the Figure we can observe residues such as I10, G13, F82 have more conservation that is 100 percent conservation. Below that V18, A31, K33, V64, N131, L134 and D147 are seen. Here we can also say that F146, K89, G147 and A149 have lowest conformation.
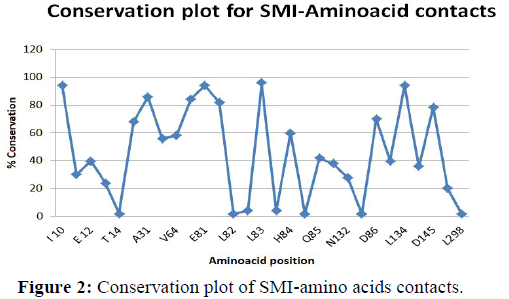
Conservation plot for amino acids interactions CDK2- inhibitor complexes
In Figure 2 conservation of CDK2 inhibitor complexes is shown. The amino acids that interact with inhibitor complexes are taken in the x-axis and percentage of conservation is taken as Y-axis [37]. Graph has been drawn using the Table 6. Here I10, E81, L83, L134 have highest value that is highest 100. A31, F80, F82, and D145 has nearer to the value 80 and T14, L82, H82, V83, T89, K88, L298 have lowest values nearer to zero.
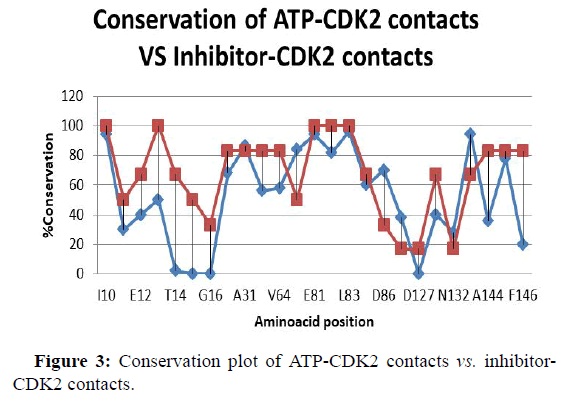
Conservation plot of ATP-CDK2 contacts vs. inhibitor CDK2 contacts
In Figure 3 conservation of ATP-CDK2 complexes vs. CDK2- inhibitor complexes is shown. The amino acids that interact with ATP and SMI complexes are taken in the X-axis and percentage of conservation is taken as y-axis. Here two values that is conservation of ATP-CDK values combined with conservation of SMI CDK values. Here we can observe that in both case I10, G13, E81, F81, F82 have maximum values [38]. In ATP-CDK lowest value is H84 and A144. But in SMI- CDK T14, Y15, G16, and D127 have lowest values. In ATP-CDK maximum value is 100 but in SMI-CDK maximum value is 94.
Ligand-protein interactions in CDK2-ATP complexes
First we considered the interactions of the natural ligand ATP in different CDK2-ATP complexes using the software in the PDB. Using this tool number of hydrogen bonds, hydrophobic bonds, hydrophilic bonds and the other bonds are found and shown in Table 8. Here we can see that hydrogen bonds present in five molecules out of six. Hydrophobic bonds, hydrophilic bonds and the other bonds are present in all six complexes [39]. In this Table interactions of 1B38 are seen to similar to 1B39 only hydrogen bond is different but we see the total interactions of these molecules it seems to be almost similar. In the case of 1FIN and 1FQ1 number of bonds seems to be different but when we compare the total interactions it is almost similar.
| Dependent variables | No. of BHB | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total |
|---|---|---|---|---|---|
| IC50 | 0.034802 | 0.391765 | 0. 13696237 | 0.090143 | -0.0096 |
| LogIC50 | 0.030922 | -0.69087 | 0.060849 | -0.08489 | -0.03881 |
| 1/IC50 | 0. 145977 | 0.666331 | 0.028395 | 0.214466 | 0.245536 |
Table 8: Single correlation coefficients with IC50, Log IC50 and 1/IC50.
Ligand protein interactions in CDK2-small molecule inhibitor complexes
Here we analyzed the various ligand protein interactions in CDK2- Small molecule inhibitor complexes. 50 CDK 2 small molecule inhibitor were analyzed and the results are shown in Table 9. Here we can see all the molecule such as 1G5S, 1GIH, 1H 1P 1PF8 hydrogen bond is absent. Hydrophilic bonds are absent in molecules such as 1H 1P, 1OI9, 1OIU, and 1OIY. Hydrophobic bonds are absent in molecules such as 1HIP, 1O 19 1OIU, and 1OIY but 1H 1P all these three bonds are absent [40].
| PDBID | IC50 | LogIC50 | 1/C | No. of BHB | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total |
|---|---|---|---|---|---|---|---|---|
| 1E1V | 17000 | 4.230449 | 5.88E-05 | 20 | 1 | 16 | 24 | 61 |
| 1E1X | 2200 | 3.342423 | 0.000455 | 28 | 3 | 10 | 20 | 61 |
| 1GII | 78 | 1.892095 | 0.012821 | 11 | 2 | 13 | 23 | 49 |
| 1GIJ | 25000 | 4.39794 | 0.00004 | 28 | 2 | 9 | 29 | 68 |
| 1H00 | 38000 | 4.579784 | 2.63E-05 | 28 | 1 | 18 | 32 | 79 |
| 1H01 | 1000 | 3 | 0.001 | 60 | 1 | 21 | 26 | 108 |
| 1H1Q | 970 | 2.986772 | 0.001031 | 7 | 1 | 22 | 28 | 58 |
| 1H1R | 2300 | 3.361728 | 0.000435 | 21 | 1 | 34 | 37 | 93 |
| 1H1S | 5.4 | 0.732394 | 0. 185185 | 45 | 4 | 16 | 36 | 101 |
| 1H07 | 3000 | 3.477121 | 0.000333 | 45 | 4 | 16 | 36 | 101 |
| 1H08 | 300 | 2.477121 | 0.00333 | 50 | 4 | 14 | 27 | 95 |
| 1KE5 | 560 | 2.748188 | 0.001786 | 27 | 2 | 16 | 24 | 69 |
| 1KE6 | 5.7 | 0.755875 | 0. 175439 | 20 | 4 | 16 | 31 | 71 |
| 1KE7 | 8.9 | 0.94939 | 0. 11236 | 12 | 5 | 10 | 36 | 63 |
| 1KE8 | 1000 | 3 | 0.001 | 13 | 3 | 22 | 31 | 69 |
| 1KE9 | 660 | 2.819544 | 0.001515 | 15 | 2 | 14 | 10 | 41 |
| 1OGU | 34 | 1.531479 | 0.029412 | 21 | 1 | 16 | 32 | 70 |
| 1OIR | 32 | 1.50515 | 0.03125 | 18 | 3 | 11 | 19 | 51 |
| 1OIT | 3 | 0.477121 | 0.333333 | 36 | 4 | 20 | 32 | 92 |
| 1P2A | 12 | 1.079181 | 0.083333 | 9 | 5 | 18 | 20 | 52 |
| 1PYE | 324 | 2.510545 | 0.003086 | 11 | 2 | 26 | 33 | 72 |
| 1R78 | 3 | 0.477121 | 0.333333 | 4 | 6 | 20 | 24 | 54 |
| 1URW | 3 | 0.477121 | 0.333333 | 57 | 4 | 17 | 33 | 111 |
Table 9: Multiple regression analysis tables.
Single correlation coefficients with IC50, log IC50 and 1/IC50
23 small inhibitors of kinase inhibitors that have IC50 values have been selected to compute single correlation coefficient. Correlation coefficient is generally used to find the relation between the two molecules [41]. It is of two types positive and negative. Here correlation analysis of IC50, log IC50 and 1/IC50 was carried out. Number of brigdes hydrogen bonds, number of hydrophilic bonds, number of hydrophobic bonds, other bonds and total number of interactions are selected as dependent variables. First correlation coefficient of IC50 with various combinations such as number of bridged hydrogen bonds, number of hydrophilic bonds, number of hydrophobic bonds, other interactions and total of these interactions has found. In case of dependent variables with number of hydrogen bonds seen that two values got almost similar values with IC50 and log IC50 but in case of 1/IC50 is different and is seen as greater than other values [42]. In case of dependent variables with number of hydrophilic bonds are seen that two values are negative that is we can say that it is a negative correlation and the other value is positive and we can say as positive correlation. So again here also 1/IC50 values with various combinations got greater value. In case of dependent variables with number of hydrophobic bonds, it is seen that one is negative correlation other two is positive correlation. Here also 1/IC50 values with various combinations got high value compared with other variables [43]. In case of dependent variables with other interactions IC50 and 1/IC50 got positive correlation but log IC50 got negative correlation [44]. Here also 1/IC50 values show greater correlation. Then correlation coefficients of total of these interactions were analyzed. Here first two values show negative correlation and third one showing positive here also 1/IC50 values with various combinations got greater value compared to IC50 and Log IC50 values. In general we can say single correlation is high in case of 1/IC50 compared to IC50 and log IC50.
Multiple regression analysis
Multiple regression and Pearson coefficients were done using 23 CDK inhibitors that have IC50 values. Number of bridges hydrogen bonds, number of hydrophilic bonds, number of hydrophobic bonds, other bonds and total number of interactions are selected as dependent variables [45]. Log values and reciprocal of IC50 values were calculated. Relationship between five sets of parameters and the IC50 values, log IC50 values and 1/IC50 were analyzed by computing correlation coefficients and by multiple regression analysis. The 23 CDK inhibitors that have been used for multiple regression analysis is shown in Table 10.
| Various combination | Correlation coefficient |
|---|---|
| 12 | 0.393499 |
| 13 | 0. 136962137 |
| 14 | 0.090143 |
| 15 | 0.009601 |
| 123 | 0.462561 |
| 124 | 0.090143 |
| 125 | 0.009601 |
| 135 | 0.009601 |
| 145 | 0. 129561752 |
| 1234 | 0.522186 |
| 1235 | 0.009601 |
| 1245 | 0. 129561752 |
| 1345 | 0.21842 |
| 12345 | 0.522186 |
Table 10: Regression analysis table with IC50.
• Number of hydrogen bonds.
• Number of hydrophilic bonds.
• Number of hydrophobic bonds.
• Other bonds.
• Total.
Multiple regression analysis of small molecule inhibitor interaction vs. biological activity
Multiple analysis of the role of the different interactions such as IC50, Log IC50 and 1/IC50 vs. biological activity was carried out. At a given time two or more interactions were considered together. The results are given in Table 11. When two interactions were considered number of hydrogen bonds and number of hydrophobic bonds gave the maximum correlation of 0.39. The above procedure was repeated for log IC50 and 1/IC50 and the results are precised in Table 12.
| Various combination | Correlation coefficient |
|---|---|
| 12 | 0.701756 |
| 13 | 0.060185 |
| 14 | 0.084893 |
| 15 | 0.038813 |
| 123 | 0.701756 |
| 124 | 0.084893 |
| 125 | 0.038813 |
| 135 | 0.038813 |
| 145 | 0.088108 |
| 1234 | 0.70176 |
| 1245 | 0.088108 |
| 1345 | 0. 131050295 |
| 12345 | 0.70176 |
Table 11: Regression analysis table with LogIC50.
• Number of hydrogen bonds.
• Number of hydrophilic bonds.
• Number of hydrophobic bonds.
• Other bonds.
• Total.
• Number of hydrophilic bonds.
• Number of hydrophobic bonds.
• Other bonds.
• Total.
| Various combination | Correlation coefficient |
|---|---|
| 12 | 0.68138 |
| 123 | 0.715931 |
| 124 | 0.214466 |
| 125 | 0.245536 |
| 145 | 0.254938 |
| 1234 | 0.717425 |
| 1235 | 0.245536 |
| 1245 | 0.254938 |
| 1345 | 0.263815 |
| 12345 | 0.717425 |
Table 12: Regression analysis table with 1/IC50.
Prediction of IC50 values based on interactions
We have made an attempt to predict the IC50 value using the concept on interaction. We setup regression equations for the 23 small molecule inhibitor complexes with IC50 value and interaction obtained with minimum distance of separation respectively. Aback check test was carried out to verify the self-consistency of the analysis; it entails calculating coefficients of multiple regressions using 23 small molecule inhibitor and computing their active IC50 values by resubstituting the values. Here calculation is done using log IC50 and 1/IC50 for good results [46]. The calculated predictive value and the log IC50 values are given in the Table 13. We found an agreement between log IC50 values and predictive value experimental observation as seen in Figure 4. From the results it is observed that the predictive value is compared with the IC50 value which gives similarity value more or less than 1. 1E1X, 1E1V, 1GII, 1GIJ, 1H08 are showing less than 1. 1KE5, 1KE9 and 1PYE show similar values. 1OGU and 1H07 are showing difference greater than 1.
| PDBID | IC50 | LogIC50 | No. of BHB | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total | Back check |
|---|---|---|---|---|---|---|---|---|
| 1E1V | 17000 | 4.230449 | 20 | 1 | 16 | 24 | 61 | 3.461787 |
| 1E1X | 2200 | 3.342423 | 28 | 3 | 10 | 20 | 61 | 2.401971 |
| 1GII | 78 | 1.892095 | 11 | 2 | 13 | 23 | 49 | 2.905079 |
| 1GIJ | 25000 | 4.39794 | 28 | 2 | 9 | 29 | 68 | 3.050602 |
| 1H00 | 38000 | 4.579784 | 28 | 1 | 18 | 32 | 79 | 3.416404 |
| 1H01 | 1000 | 3 | 60 | 1 | 21 | 26 | 108 | 3.398465 |
| 1H1Q | 970 | 2.986772 | 7 | 1 | 22 | 28 | 58 | 3.26172 |
| 1H1R | 2300 | 3.361728 | 21 | 1 | 34 | 37 | 93 | 2.941842 |
| 1H1S | 5.4 | 0.732394 | 45 | 4 | 16 | 36 | 101 | 1.63112 |
| 1H07 | 3000 | 3.477121 | 45 | 4 | 16 | 36 | 101 | 1.63112 |
| 1H08 | 300 | 2.477121 | 50 | 4 | 14 | 27 | 95 | 1.703302 |
| 1KE5 | 560 | 2.748188 | 27 | 2 | 16 | 24 | 69 | 2.851035 |
| 1KE6 | 5.7 | 0.755875 | 20 | 4 | 16 | 31 | 71 | 1.583326 |
| 1KE7 | 8.9 | 0.94939 | 12 | 5 | 10 | 36 | 63 | 1. 111215 |
| 1KE8 | 1000 | 3 | 13 | 3 | 22 | 31 | 69 | 2.022434 |
| 1KE9 | 660 | 2.819544 | 15 | 2 | 14 | 10 | 41 | 2.891583 |
| 1OGU | 34 | 1.531479 | 21 | 1 | 16 | 32 | 70 | 3.459479 |
| 1OIR | 32 | 1.50515 | 18 | 3 | 11 | 19 | 51 | 2.353705 |
| 1OIT | 3 | 0.477121 | 36 | 4 | 20 | 32 | 92 | 1.500674 |
| 1P2A | 12 | 1.079181 | 9 | 5 | 18 | 20 | 52 | 0.884951 |
| 1PYE | 324 | 2.510545 | 11 | 2 | 26 | 33 | 72 | 2.527774 |
| 1R78 | 3 | 0.477121 | 4 | 6 | 20 | 24 | 54 | 0. 190581 |
| 1URW | 3 | 0.477121 | 57 | 4 | 17 | 33 | 111 | 1.628374 |
Table 13: Predictive value table of log IC50.
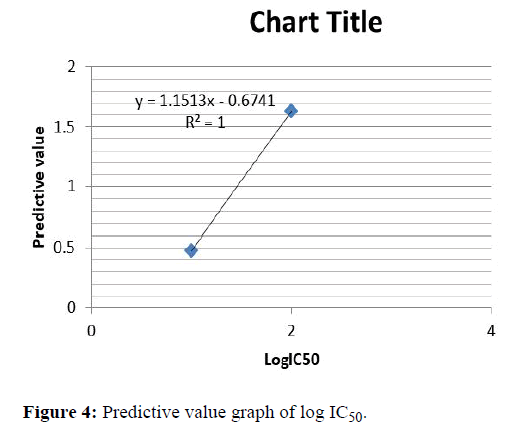
Predictive value graph for logIC50
Here predictive values are taken as X-axis and Log IC50 values are taken as Y-axis. Here 1R78, 1P2A, 1KE7, 1PYE, 1KE5, 1KE9, 1H1Q, 1H1R are matching. When comparing the LogIC50 values of these molecules it is seen that the activity values are almost similar. Using these values straight line graphs have been drawn as in Figure 4.
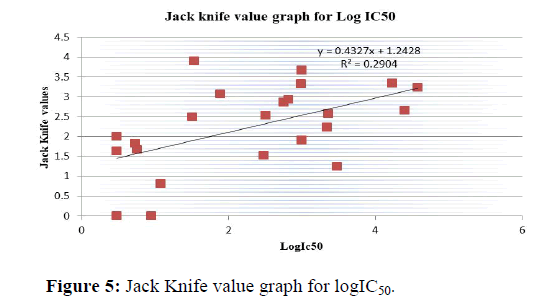
Jack knife test
We have also performed the jack knife test (leave one out of rule) for all those 23 complexes to examine the validity of the present method and the results and are included in Table 14. Third test validate the present method by determining the coefficients of multiple regression (n-1) data and then computing the IC50 values of the omitted complex. We found an agreement between the IC50 values and Jack knife value plotted in Figure 5.
| PDBID | IC50 | LogIC50 | No. of BHB | No. of hydrophilic bonds | No. of hydrophobic bonds | Other | Total | JKV |
|---|---|---|---|---|---|---|---|---|
| 1E1V | 17000 | 4.230449 | 20 | 1 | 16 | 24 | 61 | 3.346952 |
| 1E1X | 2200 | 3.342423 | 28 | 3 | 10 | 20 | 61 | 2.237571 |
| 1GII | 78 | 1.892095 | 11 | 2 | 13 | 23 | 49 | 3.06734 |
| 1GIJ | 25000 | 4.39794 | 28 | 2 | 9 | 29 | 68 | 2.660123 |
| 1H00 | 38000 | 4.579784 | 28 | 1 | 18 | 32 | 79 | 3.226579 |
| 1H01 | 1000 | 3 | 60 | 1 | 21 | 26 | 108 | 3.666906 |
| 1H1Q | 970 | 2.986772 | 7 | 1 | 22 | 28 | 58 | 3.321855 |
| 1H1R | 2300 | 3.361728 | 21 | 1 | 34 | 37 | 93 | 2.57461 |
| 1H1S | 5.4 | 0.732394 | 45 | 4 | 16 | 36 | 101 | 1.818512 |
| 1H07 | 3000 | 3.477121 | 45 | 4 | 16 | 36 | 101 | 1.246213 |
| 1H08 | 300 | 2.477121 | 50 | 4 | 14 | 27 | 95 | 1.518566 |
| 1KE5 | 560 | 2.748188 | 27 | 2 | 16 | 24 | 69 | 2.859292 |
| 1KE6 | 5.7 | 0.755875 | 20 | 4 | 16 | 31 | 71 | 1.66765 |
| 1KE7 | 8.9 | 0.94939 | 12 | 5 | 10 | 36 | 63 | 1.224553 |
| 1KE8 | 1000 | 3 | 13 | 3 | 22 | 31 | 69 | 1.902066 |
| 1KE9 | 660 | 2.819544 | 15 | 2 | 14 | 10 | 41 | 2.935676 |
| 1OGU | 34 | 1.531479 | 21 | 1 | 16 | 32 | 70 | 3.901542 |
| 1OIR | 32 | 1.50515 | 18 | 3 | 11 | 19 | 51 | 2.498339 |
| 1OIT | 3 | 0.477121 | 36 | 4 | 20 | 32 | 92 | 1.636851 |
| 1P2A | 12 | 1.079181 | 9 | 5 | 18 | 20 | 52 | 0.810898 |
| 1PYE | 324 | 2.510545 | 11 | 2 | 26 | 33 | 72 | 2.531933 |
| 1R78 | 3 | 0.477121 | 4 | 6 | 20 | 24 | 54 | 0.002181 |
| 1URW | 3 | 0.477121 | 57 | 4 | 17 | 33 | 111 | 2.006258 |
Table 14: Jack Knife test value table of LogIC50.
From the result we can observe that the similarities between the Jack knife value and IC50 value are more or less than 2. In Table 15 1R78, 1URW, 1OIT, 1OGU, 1H07, 1H08, and 1GII are showing more two differences. 1H01, 1H1Q, 1KE9, 1P2A, 1PYE, 1OIR etc. are showing less two differences.
| PDBID | LogIC50 | 1/C | Hydrophobic residues | Neutral residues | Negatively charged residues | Positively charged residues |
|---|---|---|---|---|---|---|
| 1E1V | 4.230449 | 20 | 8 | 4 | 3 | 1 |
| 1E1X | 3.342423 | 28 | 8 | 1 | 3 | 1 |
| 1GII | 1.892095 | 11 | 9 | 1 | 3 | 2 |
| 1GIJ | 4.39794 | 28 | 6 | 1 | 3 | 2 |
| 1H00 | 4.579784 | 28 | 6 | 1 | 3 | 3 |
| 1H01 | 3 | 60 | 5 | 1 | 4 | 2 |
| 1H1Q | 2.986772 | 7 | 8 | 2 | 4 | 2 |
| 1H1R | 3.361728 | 21 | 7 | 3 | 4 | 2 |
| 1H1S | 0.732394 | 45 | 9 | 3 | 3 | 2 |
| 1H07 | 3.477121 | 45 | 9 | 2 | 3 | 2 |
| 1H08 | 2.477121 | 50 | 8 | 1 | 2 | 1 |
| 1KE5 | 2.748188 | 27 | 6 | 2 | 1 | 2 |
| 1KE6 | 0.755875 | 20 | 10 | 1 | 3 | 2 |
| 1KE7 | 0.94939 | 12 | 10 | 2 | 3 | 3 |
| 1KE8 | 3 | 13 | 9 | 1 | 4 | 2 |
| 1KE9 | 2.819544 | 15 | 9 | 0 | 2 | 2 |
| 1OGU | 1.531479 | 21 | 8 | 2 | 4 | 2 |
| 1OIR | 1.50515 | 18 | 6 | 2 | 3 | 2 |
| 1OIT | 0.477121 | 36 | 6 | 1 | 3 | 1 |
| 1P2A | 1.079181 | 9 | 7 | 1 | 2 | 1 |
| 1PYE | 2.510545 | 11 | 7 | 2 | 2 | 1 |
| 1R78 | 0.477121 | 4 | 9 | 4 | 3 | 0 |
| 1URW | 0.477121 | 57 | 7 | 1 | 3 | 2 |
Table 15: Multiple regression analysis for the Log IC50 and 1/IC50.
Multiple regression analysis of aminoacid ligand interaction
Representation of aminoacid ligand interaction in small molecule inhibitor complexes: From the Table 16 the amino acid ligand interactions has been calculated from the contact map by using the hydrophobic bonds, neutral, positive charge and negative charge.
Here we can observe that the hydrophobic interactions are more in the case of 1KE6 and 1KE7 and less in the case of 1H01. Neutral residues are more in the case of 1E1V and 1R78 and in the case of 1KE9 it is absent. Negatively charged residues are almost equal in all the complexes except in case of 1KE5. Positively charged residues are high in case of 1H00 and 1R78 it is absent.
| Single regression analysis result | ||||
| Dependent variables | Hydrophobic residues | Neutral residues | Negatively charged residues | Positively charged residues |
|---|---|---|---|---|
| LogIC50 | -0.28809 | -0.05182 | 0.080357 | 0.211428 |
| 1/IC50 | 0.123239 | 0.154196 | 0.004804 | -0.30999 |
Table 16: Single regression result of amino acid ligand interaction.
Multiple regression result of amino acid ligand interaction
Here we did the multiple regression analysis for the log IC50 and 1/IC50 values. Here log IC50 showing maximum correlation correlation (0.366175) than the 1/IC50 (0.341989) but almost similar (Table 17).
| Multiple regression analysis result | |
|---|---|
| Dependent variables | |
| LogIC50 | 0.366175 |
| 1/IC50 | 0.341989 |
Table 17: Multiple regression result of amino acid ligand interaction.
Conclusion
Understanding the structural basis of small molecular ligand binding to enzyme can pave way for design of novel inhibitors, lead modification and eventually in structure based drug discovery. Keeping this in mind, in the present work we have analyzed 50 cyclin dependent kinase (which has a key role in cell signaling) small molecule inhibitor complexes and 6 CDK2-ATP complexes. 23 CDK2 small molecule inhibitor complexes that have IC50 values out of 50 CDK2 inhibitor complexes are also taken for various types of analysis. As binding to the ATP site and biological activity may be dependent on the different non-covalent interactions such as hydrogen bonds, hydrophobic bonds hydrophilic bonds and electrostatic vaanderwaals such as other interactions. Here we have analyzed these interactions in the CDK2-ATP complexes as well as the CDK2 inhibitor complexes. The 2D and 3D similarity analysis of 23 CDK2 small molecule inhibitor that have IC50 values enabled us to relate structural similarity with biological activity. Further from various noncovalent interactions we have developed multiple regression models to accounts for the experimentally observed IC50 values and to predict biological activity based on the different non-covalent interactions.
References
- Zhang J, Yang PL, Gray NS (2009) Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9:28-39.
[Crossref] [Google Scholar] [PubMed]
- Malumbres M (2014) Cyclin dependent kinases. Genome Biol 15:122-132.
[Crossref] [Google Scholar] [PubMed]
- Morgan DO (2007) The cell cycle: Principles of control. New Sci Press, London.
- Lee MG, Nurse P (1987) Complementation used to clone a human homologue of the fission yeast cell cycle control gene CDC2. Nature 327:31-35.
[Crossref] [Google Scholar] [PubMed]
- Knight ZA, Shokat KM (2005) Features of selective kinase inhibitors. Chem Biol 12:621-637.
[Crossref] [Google Scholar] [PubMed]
- Card GL, Knowles P, Laman H, Jones N, McDonald NQ (2000) Crystal structure of a gamma-Herpesvirus cyclin-cdk complex. EMBO J 19:2877-2888.
[Crossref] [Google Scholar] [PubMed]
- Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, et al. (2005) The structure of cyclin E1/CDK2: Implications for CDK2 activation and CDK2-independent roles. EMBO J 24:452-463.
[Crossref] [Google Scholar] [PubMed]
- Lucking U, Siemeister G, Schafer M, Briem H, Kruger M, et al. (2007) Macrocyclic aminopyrimidines as multi target CDK and VEGF-R inhibitors with potent anti-proliferative activities. ChemMedChem 2:63-77.
[Crossref] [Google Scholar] [PubMed]
- Watson JD, Baker T, Bell SP, Gann A, Levine M, et al. (2008) Molecular biology of the gene. 6th Edition, Cold spring harbor laboratory press, New York.
- Morgan DO (1997) Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13:261-291.
[Crossref] [Google Scholar] [PubMed]
- Lew J (2003) MAP kinases and CDKs: Kinetic basis for catalytic activation. Biochem 42:849-856.
[Crossref] [Google Scholar] [PubMed]
- Avendano C, Menendez JC (2015) Medicinal chemistry of anticancer drugs. Elsevier. 1-5.
- Finn RS, Dering J, Conklin D, Kalous O, Cohen DJ, et al. (2009) PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11:1-13.
[Crossref] [Google Scholar] [PubMed]
- Berman H, Henrick K, Nakamura H, Markley JL (2007) The worldwide Protein Data Bank (wwPDB): Ensuring a single, uniform archive of PDB data. Nucleic Acids Res 35:D301- D303.
[Crossref] [Google Scholar] [PubMed]
- Moreland JL, Gramada A, Buzko OV, Zhang Q, Bourne PE (2005) The Molecular Biology Toolkit (MBT): A modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21.
[Crossref] [Google Scholar] [PubMed]
- Michalsky E, Dunkel M, Goede A, Preissner R (2005) Super ligands-a database of ligand structures derived from the protein data bank. BMC Bioinformatics 6:1-6.
[Crossref] [Google Scholar] [PubMed]
- Sridhar J, Akula N, Pattabiraman N (2006) Selectivity and potency of cyclin dependent kinase inhibitors. AAPS J 8:E204- E221.
[Crossref] [Google Scholar] [PubMed]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK (2007) BindingDB: A web accessible database of experimentally determined protein-ligand binding affinities. Nucleic Acids Res 35:D198- D201.
[Crossref] [Google Scholar] [PubMed]
- Brown NR, Noble ME, Lawrie AM, Morris MC, Tunnah P, et al. (1999) Effects of phosphorylation of threonine 160 on cyclin-dependent kinase 2 structure and activity. J Biol Chem 274:8746-8756.
[Crossref] [Google Scholar] [PubMed]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, et al. (1995) Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376:313-320.
[Crossref] [Google Scholar] [PubMed]
- Song H, Hanlon N, Brown NR, Noble ME, Johnson LN, et al. (2001) Phosphoprotein-protein interactions revealed by the crystal structure of kinase associated phosphatase in complex with phosphoCDK2. Mol Cell 7:615-626.
[Crossref] [Google Scholar] [PubMed]
- Schulze-Gahmen U, De Bondt HL, Kim SH (1996) High resolution crystal structures of human cyclin dependent kinase 2 with and without ATP: Bound waters and natural ligand as guides for inhibitor design. J Med Chem 39:4540-4546.
[Crossref] [Google Scholar] [PubMed]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP (1996) Crystal structure of the p27Kip1 cyclin dependent kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331.
[Crossref] [Google Scholar] [PubMed]
- Lawrie AM, Noble ME, Tunnah P, Brown NR, Johnson LN, et al. (1997) Protein kinase inhibition by staurosporine revealed in details of the molecular interaction with CDK2. Nat Struct Biol 4:796-801.
[Crossref] [Google Scholar] [PubMed]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, et al. (1998) Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science 281:533-538.
[Crossref] [Google Scholar] [PubMed]
- Shewchuk L, Hassell A, Wisely B, Rocque W, Holmes W, et al. (2000) Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase. J Med Chem 43:133-138.
[Crossref] [Google Scholar] [PubMed]
- Wang S, Meades C, Wood G, Osnowski A, Anderson S, et al. 2-Anilino-4-(thiazol-5-yl) pyrimidine CDK inhibitors: Synthesis, SAR analysis, X-ray crystallography, and biological activity. J Med Chem. 47:1662-1675.
[Crossref] [Google Scholar] [PubMed]
- Arris CE, Boyle FT, Calvert AH, Curtin NJ, Endicott JA, et al. (2000) Identification of novel purine and pyrimidine cyclin dependent kinase inhibitors with distinct molecular interactions and tumor cell growth inhibition profiles. J Med Chem 243:2797-2804.
[Crossref] [Google Scholar] [PubMed]
- Davies TG, Tunnah P, Meijer L, Marko D, Eisenbrand G, et al. (2001) Inhibitor binding to active and inactive CDK2: The crystal structure of CDK2-cyclin A/indirubin-5-sulphonate. Structure 9:389-397.
[Crossref] [Google Scholar] [PubMed]
- Davis ST, Benson BG, Bramson HN, Chapman DE, Dickerson SH, et al. (2001) Prevention of chemotherapy induced alopecia in rats by CDK inhibitors. Science 291:134-137.
[Crossref] [Google Scholar] [PubMed]
- Dreyer MK, Borcherding DR, Dumont JA, Peet NP, Tsay JT, et al. (2001) Crystal structure of human cyclin dependent kinase 2 in complex with the adenine derived inhibitor H717. J Med Chem 44:524-530.
[Crossref] [Google Scholar] [PubMed]
- Ikuta M, Kamata K, Fukasawa K, Honma T, Machida T, et al. (2001) Crystallographic approach to identification of Cyclin Dependent Kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein. J Biol Chem 276:27548-27554.
[Crossref] [Google Scholar] [PubMed]
- Gibson AE, Arris CE, Bentley J, Boyle FT, Curtin NJ, et al. (2002) Probing the ATP ribose binding domain of cyclin-dependent kinases 1 and 2 with O (6)-substituted guanine derivatives. J Med Chem 45:3381-3393.
[Crossref] [Google Scholar] [PubMed]
- Davies TG, Bentley J, Arris CE, Boyle FT, Curtin NJ, et al. (2002) Structure based design of a potent purine based cyclin-dependent kinase inhibitor. Nat Struct Biol 9:745-749.
[Crossref] [Google Scholar] [PubMed]
- Beattie JF, Breault GA, Ellston RP, Green S, Jewsbury PJ, et al. (2003) Cyclin dependent kinase 4 inhibitors as a treatment for cancer. Part 1: Identification and optimisation of substituted 4, 6-bis anilino pyrimidines. Bioorg Med Chem Lett 13:2955-2960.
[Crossref] [Google Scholar] [PubMed].
- Bramson HN, Corona J, Davis ST, Dickerson SH, Edelstein M, et al. (2001) Oxindole based inhibitors of Cyclin-Dependent Kinase 2 (CDK2): Design, synthesis, enzymatic activities, and X-ray crystallographic analysis. J Med Chem 44:4339-4358.
[Crossref] [Google Scholar] [PubMed]
- Sayle KL, Bentley J, Boyle FT, Calvert AH, Cheng Y, et al. (2003) Structure based design of 2-arylamino-4-cyclohexylmethyl-5-nitroso-6-aminopyrimidine inhibitors of cyclin-dependent kinases 1 and 2. Bioorg Med Chem Lett 13:3079-3082.
[Crossref] [Google Scholar] [PubMed]
- Hardcastle IR, Arris CE, Bentley J, Boyle FT, Chen Y, et al. (2004) N2-substituted O6-cyclohexylmethylguanine derivatives: Potent inhibitors of cyclin dependent kinases 1 and 2. J Med Chem 47:3710-3722.
[Crossref] [Google Scholar] [PubMed]
- Anderson M, Beattie JF, Breault GA, Breed J, Byth KF, et al. (2003) Imidazo 1, 2-a pyridines: A potent and selective class of cyclin dependent kinase inhibitors identified through structure based hybridisation. Bioorg Med Chem Lett 13:3021-3026.
[Crossref] [Google Scholar] [PubMed]
- Liu JJ, Dermatakis A, Lukacs C, Konzelmann F, Chen Y, et al. (2003) 3, 5, 6-trisubstituted naphthostyrils as CDK2 inhibitors. Bioorg Med Chem Lett 13:2465-2468.
[Crossref] [Google Scholar] [PubMed]
- Moshinsky DJ, Bellamacina CR, Boisvert DC, Huang P, Hui T, et al. (2003) SU9516: Biochemical analysis of cdk inhibition and crystal structure in complex with cdk2. Biochem Biophys Res Commun 310:1026-1031.
[Crossref] [Google Scholar] [PubMed]
- Johnson LN, de Moliner E, Brown NR, Song H, Barford D, wt al. (2002) Structural studies with inhibitors of the cell cycle regulatory kinase cyclin-dependent protein kinase 2. Pharmacol Ther 93:113-124.
[Crossref] [Google Scholar] [PubMed]
- Wu SY, McNae I, Kontopidis G, McClue SJ, McInnes C, et al. (2003) Discovery of a novel family of CDK inhibitors with the program LIDAEUS: Structural basis for ligand induced disordering of the activation loop. Structure 11:399-410.
[Crossref] [Google Scholar] [PubMed]
- Hamdouchi C, Keyser H, Collins E, Jaramillo C, De Diego JE, et al. (2004) The discovery of a new structural class of cyclin-dependent kinase inhibitors, aminoimidazo 1, 2-a pyridines. Mol Cancer Ther 3:1-9.
[Google Scholar] [PubMed]
- Luk KC, Simcox ME, Schutt A, Rowan K, Thompson T, et al. (2004) A new series of potent oxindole inhibitors of CDK2. Bioorg Med Chem Lett 14:913-917.
[Crossref] [Google Scholar] [PubMed]
- Byth KF, Cooper N, Culshaw JD, Heaton DW, Oakes SE, et al. (2004) Imidazo 1,2-b pyridazines: A potent and selective class of cyclin dependent kinase inhibitors. Bioorg Med Chem Lett 14:2249-2252.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi