Research Article, J Appl Bioinforma Comput Biol Vol: 9 Issue: 6
Structural Arrangement of Q_A Binding Site from the Rhodobacter Sphaeroides Photosynthesis Reaction Center
Madhu Sudhan Bhusal1*, Kiran Pudasainee1 and Hari Prasad Lamichhane1,2
1Deprtment of Physics, St. Xavier’s College, Maitighar, Kathmandu, Nepal
2Central Department of Physic s, Tribhuvan University, Kirtipur, Kathmandu, Nepal
*Corresponding Author: Madhu Sudhan Bhusal
Department of Physics, St. Xavier’s College, Maitighar, Kathmandu, Nepal
E-mail: binalmsb@gmail.com
Received: September 12, 2020 Accepted: December 10, 2020 Published: December 17, 2020
Citation: Bhusal MS, Pudasainee K, Lamichhane HP (2020) Structural Arrangement of QA Binding Site from the Rhodobacter Sphaeroides Photosynthesis Reaction Center. J Appl Bioinforma Comput Biol 9:6. doi: 10.37532/jabcb.2020.9(6).186
Abstract
The Photochemical reaction center in purple bacteria is carried out in the pigments which are bound and arranged in specific way by the proteins. In this work, we construct the potential energy surface from the Q_A binding site at the periphery of 10 Å sphere, to make more visualize the structural arrangement of Q_A binding site. The ubiquinone (Q_A) molecule contains two carbonyl oxygen atoms in the benzoquinone ring. The O_1 and O_4 atoms of ubiquinone make hydrogen bonds with ALA260 N - O_1 (≈2.84 Å away) and HIS219 N - O_4 (≈2.78 Å away) respectively, which help to stabilize the ubiquinone. The rest of the molecule is surrounded by mostly hydrophobic residues making van der Waal’s contacts. These segments form the entrance of the Q_A binding pocket and membrane binding surface. In fact, the Q_A binding site is deeply buried in the protein complex, and the positively charged residues surrounded near the membrane involved in interacting with the ubiquinone head group to facilitate the electron transfer
Keywords: Potential Energy Surface; Purple Bacteria Reaction Center; Q_A binding site; Rhodobacter Sphaeroides (1AIJ)
Abbreviations
RC - Reaction Center; QA-Ubiquinone; Rb. Sphaeroides - Rhodobacter Sphaeroides; ps - picosecond.
Introduction
From the beginning of Universe, light plays an important role in the origin of life on earth. All the life on earth depends on the sun light. The light from the sun is received by plants and other organisms that have capacity of growing by the process of Photosynthesis [1]. Firstly, the light energy is absorbed by the light-harvesting antenna complexes and then transferred to the photosynthesis reaction center, where the primary charge separation and the electron transfer occurs. Photosynthetic reaction centers from purple bacteria are the best known membrane protein complexes [2]. Several integral membrane proteins and the number of co-factors are present in the photosynthesis reaction center [3]. The three dimensional structure of the reaction center is determined from the photosynthetic bacterium Rhodobacter Sphaeroides by x-ray diffraction at the resolution of 2.2 Å [4].
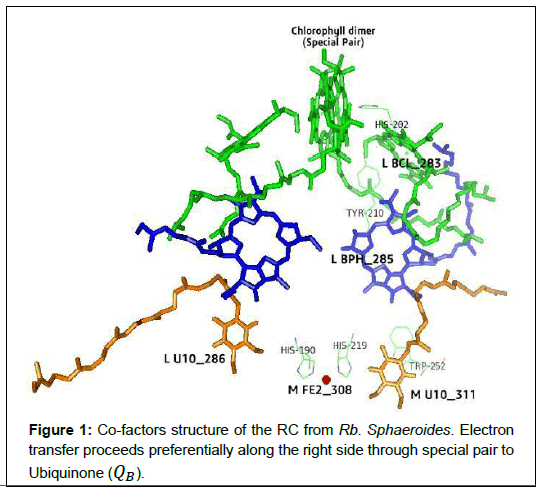
The reaction center from the Rb. Sphaeroides containing several integral membrane proteins and pigments molecules. According to their apparent molecular weight, they consists of three protein sub-units called H (Heavy), M (Medium) and L (Light), which is determined by the sodium dodecylsulphate polyacrylamide gel electrophoresis. The core of the RC complex bind by the photosynthetic pigments is formed by the L and M subunits but H subunits is frequently removed without impairing the photochemistry [2]. The L and M subunits containing two bacteriochlorophylls (D) called special pair, two bacteriochlorophylls (BchlA and BchlB) forming a monomer, two bacteriopheophytins (BPA and BPB), one non-heme iron, one ubiquinone (QA) as a primary elecron acceptor and another ubiquinone (QB) as a secondary electron acceptor. These pigment molecules are arranged in a nearby symmetrical branches (called ’A’ and ’B’) [5] as shown in Figure 1.
The three dimensional structure of the photosynthetic reaction center is a transmembrane protein which converts the light energy into chemical energy [6]. The absorbed light energy by the light harvesting antenna is transfer to the primary electron donor (D). The lowest single electronic excited state is highly reduced, and rapidly transfers an electron to BchlA in about 3 ps, and then transfer to a BPA in 1 ps. Further on the primary acceptor QAreceives the electron in about 200 ps via BPA. Finally, the electron is transferred further to the secondary quinone QB in about 6 s. The photosynthesis RC helps to charge separation at each side of the membrane, and then other chemical reactions occur [7-9]. QA and QB are chemically identical but behave differently. QA bound with the M subunit in a relatively hydrophobic pocket, only one electron acceptor at a time. But QB bound with the L subunit, surrounded by charged and polar residues, accepting two electron from QA sequentially at a time [10].The structural arrangement of pigment molecules in the purple bacterial reaction center from the Rb. Sphaeroides (1AIJ), the organism that is widely used for spectroscopic and site directed mutagensis studies of the RC [11]. Therefore in this paper, the details of the structural arrangement of QA binding site is discussed and determine the potential binding pocket of the QA binding site from the Rhodobacter Sphaeroides.
Material and Methods
ONIOM (Our own N-Layered Integrated Molecular Orbital
+ Molecular Mechanics) is the computational technique models defining the large molecules in two or three layers within the structure that are treated at different levels of accuracy. By testing a large number of ONIOM can accurately reproduce the standard CASSCF (10e/10o) (10 active electrons in 10 orbitals) results for only 10% of the computer time [12].
In this method, the entire molecular system is divided into two parts: the model system and environment system. The pigment molecules, chemically important layer is treated with the accurate QM (Quantum Mechanics) method while the environment system is calculated with less accurate but more efficient MM (Molecular Mechanics) method. Calibration studies have demonstrated the prediction result are essentially equivalent to those that would be produced by the high accuracy method alone on the entire molecule [13].
The quantum level calculation is performed by using DFT (Density Function Theory) with B3LYP functional and 6-31G basis set. The AMBER level is used to calculate the electronic embedding of the protein binding site on the quinone molecule at low level and Gaussian key word ONIOM = embed [14].
From the position (1, 4) of carbonyl oxygen, as shown in figure 2, atom manipulates the surrounded amino acids at the periphery of 10 Å sphere of QA binding site by using Swiss PDB Viewers. GAUSSIAN 05W (Frisch et al. 2004) is used to optimize the molecules and were employed for all the calculations. The PMV and PyMol is used to construct the potential energy surface of the QA binding site because they have number of customizable features and comes with many pluggable commands ranging from displaying molecular surfaces to advanced volume rendring [15].
Result and Discussion
The pigments and proteins which convert light energy into chemical energy and being the process of electron transfer are known as reaction center. In photosynthetic reaction center from the purple bacteria, ubiquinones plays an important role in biological proton, and electron transferring processes which occurs in photosynthesis [16]. In Rb. Sphaeroides purple bacterial reaction centers two ubiquinone (UQ) molecules called QA and QB, acts as terminal electron acceptors. Both ubiquinones molecules have very different redox functions, which testimonies to the flexibility of UQs in biological process because the pigment-protein interactions must modulate their functional properties [17,18].
A. General Organization of the Photosynthesis Reaction Center Co-factors
The co-factor of the photosynthetic reaction center, which plays an important role to transfer the electron from the Rb. Sphaeroides are arranged as shown in Figure 1. These co-factors are generally arranged along two branches called 2-fold symmetry axis (i.e. branch A and Branch B). Here, the line joining the center of the chlorophyll dimer and the Fe atom represents only an approximate 2-fold symmetry axis. The electron pathway is predicted from Spectroscopic measurements along the branch A. The primary donor Bchl2 (i.e. special pair) follows by Bchl2 (i.e. L BCL283) and BPhe follows upto secondary quinone (QB). The distance and angles between the cofactors are shown in Table 1 and Table 2.
| Co- factors | Angle between ring normals, (0) |
Distance between ring canters, (Å) |
|---|---|---|
| M BCL_310-L BCL_282 | 9.30 | 7.52 |
| M BCL_310-L BCL_283 | 122.24 | 12.94 |
| M BCL_310- M HIS_202 | 82.84 | 3.46 |
| M BCL_310-M TYR_210 | 68.10 | 9.97 |
| M BCL_283- M HIS_202 | 136.82 | 11.97 |
| M BCL_283- M TYR_210 | 94.41 | 5.89 |
| M HIS_ 202- M TYR_210 | 60.58 | 10.08 |
| L BCL_ 310- L BPH_285 | 71.84 | 19.52 |
| L BCL_ 283 - L BPH_285 | 65.38 | 10.83 |
| L BCL_ 283- M TYR_210 | 94.41 | 5.89 |
| M TYR_ 210- L BPH_285 | 143.99 | 9.64 |
| L BPH_ 285- M U10_ 311 | 27.51 | 13.90 |
| L BPH_ 285- M TRP_252 | 48.58 | 10.55 |
| L BPH_ 285- M PHE_258 | 71.80 | 13.52 |
| M TRP_ 252- M U10_311 | 22.07 | 5.12 |
| M PHE_258- M U10_ 311 | 63.22 | 9.25 |
| M U10_311- M FE2_ 308 | NA | 8.87 |
| M U10_311- M HIS_ 219 | 46.70 | 6.12 |
| M HIS_219 – M FE2_308 | NA | 3.26 |
| M FE2_ 308 – L U 10_286 | NA | 11.63 |
| M FE2_ 308 – L HIS_190 | NA | 3.18 |
| L HIS_190 – L U 10_ 286 | 47.41 | 8.67 |
| M U10_311- L U 10_ 286 | 8.48 | 19.64 |
Table 1: Dihedral angle and distance between purple bacteria RC and its surrounded amino acids.
| Co- factors | Angle between ring normals, (o) |
Distance between ring canters, (Å) |
|---|---|---|
| M U10_311-L PHE_005 | 94.46 | 12.30 |
| M U10_311- L HIS_190 | 39.12 | 11.50 |
| M U10_311- L HIS_230 | 48.89 | 10.38 |
| M U10_311-L PHE_285 | 27.32 | 13.90 |
| M U10_311- H TYR_040 | 149.64 | 13.58 |
| M U10_311- M PHE_216 | 137.26 | 11.68 |
| M U10_311- M HIS_219 | 133.61 | 6.12 |
| M U10_311- M PHE_251 | 99.38 | 8.69 |
| M U10_311- M TRP_252 | 158.50 | 5.12 |
| M U10_311- M TRP_254 | 133.59 | 13.26 |
| M U10_311- M PHE_258 | 62.99 | 9.25 |
| M U10_311- M HIS_266 | 123.87 | 8.87 |
| M U10_311- M TRP_268 | 14.31 | 8.97 |
Table 2: Dihedral angle and distance of 10 Å sphere of QA binding site and its amino acids.
The BChl2 Dimer: It is also called primary donor. After getting the solar energy from the sunlight, it transfer an electron to Pheophytin (BPh). In the Rb. Sphaeroides, the distance between 2-ring centers of dimer is ≈ 7.52 Å. The two BChls (dimer) are approximately parallel, and the dihedral angle between 2-ring is 9.300. The average distance between 2-ring of special pair is ≈ 3.76 Å, and are located average distance from the central Mg atoms. A 5-coordinates Mg with the acetyl group, it provides the hydrogen bonding to the nearest atom (OBD) of M HIS202 at a distance is ≈ 2.27 Å far away and the dihedral angle between them is 82.840. This hydrogen bond helps to fasten electron transfer.
The BChl Monomer: The X-ray shows that the position of BChl monomer is in between BChl2 and BPhe (Figure 1). BChl monomer and BChl dimer is 12.94 Å far from the ring center, whereas the normal angle made between them is 122.240. The function of BChl monomers is not clear yet, but it plays an important role in facilitating electron transfer from BChl2 to BPhe. In between BChl2 and BChl, M HIS202 is present. The distance between nearest atom (ND1 - OBD) of HIS_202 and BCL_283 is ≈ 5.18 Å which decrease the distance between them and helps to transfer the electron faster.
The Pheophytins (BPhe): The BPheA is located in between the BChl monomer and Quinone (QA) as shown in fig 1. One of the BPhes (i.e BPheA) serves as an intermediate acceptor, while other is not involved in electron transfer. The time required to transfer an electron from BChl2 to BPheA is ≈ 4ps. The distance between 2-ring center of BChl monomer and BPheA is ≈ 10.83 Å. In between BChl2 and BPheA, the M TYR210 is present. The distance between them is ≈ 9.97 Å and ≈ 9.64 Å respectively.
But the distance between nearest atom (CG - CAB) of BPheA to TYR210 is ≈ 4.39 Å. M TYR210 is in Van der Waals contacts with the ring of BChl2 and BPheA. It’s possible role in hydrogen bonding to M HIS202, which may help to conduct an electron transfer from BChl2 to BPheA..
The Primary Quinone (QA): The primary Quinone (QA) is considerably closer to BPheA. The time required to electron transfer proceed from BPheA to QA is ≈ 200ps. QA receive the electron from BPheA, which is separated by a distance equal to ≈ 13.90 Å from the ring center. The M TRP252 is located in between BPheA and QA. M TRP252 is ≈ 10.55 Å far away from the ring center of BPheA and ≈
5.12 Å from the QA. But the distance between nearest atom (NE1 - O5) of TRP252 and QA is ≈ 4.61 Å. The structure suggests that M TRP252 plays a likely role in the electron transfer process.
The Secondary Quinone (Qb): The secondary quinone is the final electron acceptor of the reaction center. When this quinone becomes doubly reduced in Rb. Sphaeroides the electron transfer occurs 6 μs from QA to QB [19]. The separation distance between two quinones is ≈ 19.64 Å from each ring center. The distance between QA and QB seems large for a fast electron transfer. Indeed, the gap is bridged by L HIS190 and M HIS219 which are located in between two quinones.
QA is more nearer to the M HIS 219, which is separated from each ring center is ≈ 6.12 Å. And the distance between nearest atom (O2 – ND1) of QA and HIS219 is ≈ 2.78 Å. The QB is nearer to the L HIS190, besides ring center is ≈ 8.67 Å. And the distance between nearest atom (ND1 -O4) of QB and HIS190 is ≈ 5.21 Å. These two histidines plays a role in the electron transfer from QA to QB, suggested by this arrangement. The nearest atom helps for effective overlap of a fast electron transfer.
The Nonheme Iron (Fe):
The Fe2+ is located between two quinones. In Rb. Sphaeroids it is coordinated to four histidines (L190, L230, M219 and M266). The distance between QA to Fe2+ is ≈ 8.87 Å and distance between Fe2+ to QB is ≈ 11.63 Å. From QA to QB, the electron moves with in 6 μs. The nonheme Iron, located in between QA and QB, does not seem to play an essential role in electron transfer [20].
Instead of these, at the periphery of Fe2+, HIS219 and HIS190 amino acids are located. The edge distance between such histidines are ≈ 2.18 Å and ≈ 2.08 Å respectively, which helps to transfers the electron significantly.
B. QA Binding site
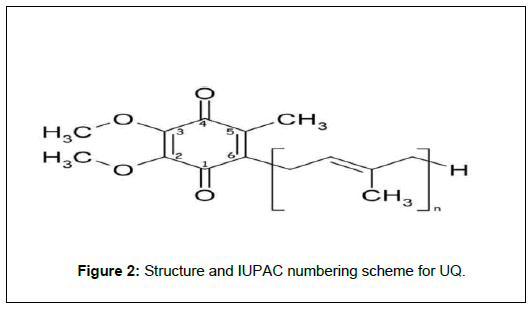
Ubiquinone is a 2, 3-dimethoxy, 5-methyl, 6-isoprenoid benzoquinone. This Figure 2, shows the structure and IUPAC (International Union of Pure and Applied Chemistry) numbering for ubiquinone.
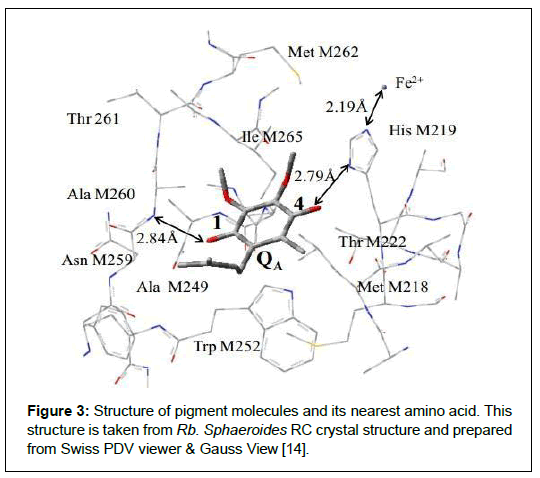
At the periphery of 10 Å QA binding site of Rb. Sphaeroides, it contains 1-Ubiquinone molecule, 50-amino acids, 6-water molecules and a non-heme iron atom. Two carbonyl groups of QA at position C1 and C4 provide hydrogen bonds to an M ALA260 and M HIS219 respectively. M ALA260 is at 2.84 Å away from the C1 = O and M HIS219 is at 2.79 Å away from the C4 = O carbonyl group. M THR222 is the possible candidate which can give H-bond to C4 = O carbonyl group. Other amino acids (M ILE265 & M MET262) are in vicinity of methoxy groups as shown in Figure 3.
Figure 3: Structure of pigment molecules and its nearest amino acid. This structure is taken from Rb. Sphaeroides RC crystal structure and prepared from Swiss PDV viewer & Gauss View [14]
C. QA Binding site
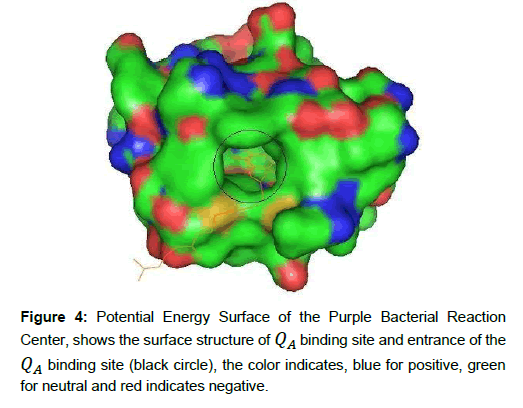
Photochemical reaction in purple bacterial RC is carried out in the pigments which are bound and arranged in specific way by the protein as shown in Figure 4 and Figure 5 respectively. In QA site, the ubiquinone (QA) contain two carbonyl oxygen atoms in the benzoquinone ring. The O2 atom of ubiquinone (QA) makes hydrogen bond to the backbone nitrogen (ND1) of HIS219 (i.e. ≈ 2.79 Å apart) and O5 atom of QA makes hydrogen bond with the backbone nitrogen (N) of ALA260 (≈ 2.84 Å apart). The rest of molecules are surrounded by mostly hydrophobic residues making van der Waal’s contacts [21]. The QA binding site is deeply buried in the protein complex, containing positively charge, negatively charge and neutral amino acids. The electron move along a chain of electron donor and electron acceptors, which is connected by hydrogen bond [11]. In QAbinding site, amino acids are located at the surface of the molecule and surrounds the ubiquinone. These segments form the entrance of the QA binding pocket and likely form the membrane binding surface. In this binding surface, the surrounding positively charged groups (blue color) probably involved to interacting with negatively charged ubiquinone, as shown in Figure 5. It is also generally believed that water binding positions were simulated and empty space was filled up with solvent using from Rb. Sphaeroides RC structures [22].
In the Photosynthesis RC, finally, we detected the potential energy surface of the purple bacterial reaction center. The QA binding site is deeply buried in the protein complex and forming the entrance of the QA binding pocket, containing positively charge, negatively charge and neutral amino acids. The surrounded positively charged groups involved to interacting with negatively charged ubiquinone.
Binding Energy
Binding Energy is the energy required to decompose a molecule, atom or nucleus into its constituent particles, equal to the energy equivalent of the mass defect. The binding energy of a QA binding site at the periphery of 10 Å has been calculated by using this formula,
EQM/MM = EQM + EMM – EQM & MM (1)
Where, EQM is the energy of Ubiquinone; EMM is the energy of the Amino Acid.
Different types of amino acids are present at the periphery of QAbinding site. To calculate the binding energy, the basis set 6-31G is used in DFT (B3LYP) level of approximation. The calculated value of binding energy was found to be 1.67eV, which is positive (+ve) value. The positive value indicates that the pigment molecules at the periphery of QAbinding site are bounded and arranged in specific way by the proteins.
It is clear that there are regions that are surface exposed with solvent accessible and holds reasonable criteria to become potential epitopes for vaccines and can be augmented by appropriate adjuvants which lead to effective protection against SARS-CoV-2 infections.
Conclusion
The conversion of light energy to chemical energy in photosynthesis is the most important biological process on earth. This conversion of energy is possible because of specific arrangement of pigments in the photosynthesis reaction center. In this study, the detected potential energy surface shows that, the QA binding site is deeply buried in the protein complex and forming the entrance of the QA binding pocket.
References
- Deisenhofer J, N(1993) The Photosynthetic Reaction Center. Academic Press San Diego.
- Roy C, Lancaster DUEHM(1995) The Structures of Photosynthetic Reaction Centers from Purple Bacteria as Re-vealed by X-Ray Crystallography. Anoxygenic Photosynthetic Bacteria 2: 503-526.
- Breton J, Vermeglio A (1992) The Photosynthetic Bac-terial Reaction Center II, NATO-ASI Series A. Life Sciences. Plenum Press, New York, 237.
- Nobel Prize.org (2020) Photosynthesis: Chemical energy from light (2020) NobelPrize.org. Nobel Media AB 2020. Sat. 14 Nov.
- Huber R (1989) A structural basis of light energy and electron transfer in biology. The EMBO Journal 8: 2125–2147.
- Ermler UM, Michel SH (1994) Structure and function of the photosynthetic reaction center from Rhodobacter sphaeroides. J Bioenerg Biomembr 26: 5–15.
- Marcus R (1997) Electron transfer reactions in chemistry. Theory and experiment. Pure Appl. Chem., 69: 13–29.
- Li J, Gilroy D, Tiede DM, Gunner, MR (1998) Biochemistry 37: 2818-2829.
- Tiede, DM, Vazquez J, Cordova J, Marone, P. A. (1996) Biochemistry 35, pp. 10763-10775
- Lavergne J, Matthews C, Ginet N (1999) Biochemistry 38: 4542-4552.
- Ermler U, Fritzbch G, S. K. B, Michel H (1994) Struc-ture of the Photosynthesic Reaction Centre from Rhodobacter Sphaeroides at 2.65 A˚ Resolution: Cofactors and Protein - Cofactor Interactions. Max Plank - Institute fur Biophysik, Germany 2:925–936.
- Vreven T, Morokuma, K (2000) The ONIOM (our own N-layered integrated molecular orbital + molecular mechanics) method for the first singlet excited (S1) state photoisomeriza-tion path of a retinal protonated Schiff base. J. Chem. Phys 113: 2969.
- Dapprich S, Komaromi I, K. S. B. K. M, Frisch, MJ (1999) A New ONIOM Implementation in Gaussian 98. 1. The Calculation of Energies, Gradients and Vibrational Frequencies and Electric Field Derivatives. J. Mol. Struct (Theochem) 462: 1–21.
- Lamichhane HP (2011) Calculated Vibrational Properties of Quinones in Photosynthetic Reaction Centers. PhD Thesis. Georgia State University, USA.
- Dapprich S (2010) Molecular Graphics Laboratory. JMGLTools Website.
- Trumpower B (1982) Function of Quinones in Energy Con-serving Systems. Academic, New York.
- Srinivasan N, Golbeck, J (2009) Protein-cofactor inter-actions in bioenergetic complexes: The role of the A1A and A1B phylloquinones in photosystem I. Biochim Biophys Acta 1787: 1057-1088.
- Wraight C, Gunner M (2009) The acceptor quinones of purple photosynthetic bacteria Structure and spectroscopy. Springer, Dordrecht, 28: 379-405.
- The Nobel Prize in Chemistry (1992) Nobel Media AB 2020. NobelPrize.org.
- Huber R, H. Michel (1988) A Structural Basis of Light Energy and Electron Transfer in Biology. Nobel Lecture.
- Zhang J, F.E.F, Kim JP (2006) Structure of ElectronTransfer Flavoprotein-Ubiquinone Oxidoreductase and Electron Transfer to the Mitochondrial Ubiquinone Pool. Department of Biochemistry, Medical College of Wisconsin, WI 53226.
- Beroza P, Fredkin DR, MYO, Feher, G (1992) Proton transfer path ways in the reaction center of Rhodobacter sphaeroides: a computational study. In The Photosynthetic Bacterial Reaction Center I: Structure, Spectroscopy and Dynamics. J.Breton, and A. Vermeglio, eds, Plenum Press, New York, 363–374.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi