Research Article, J Athl Enhanc Vol: 12 Issue: 5
DNA Methylation on PPARG and its Relation to Sports Injury and Pain in Male Football (Soccer) Players at all Levels
Christopher Collins*
1Department of Data S cience, Muhdo Health Ltd, Adastral Park, Ipswich, United Kingdom
*Corresponding Author: Christopher Collins,
Department of Data Science, Muhdo Health Ltd, Adastral Park, Ipswich, United Kingdom
E-mail: chris.collins@muhdo.com
Received date: 27 August, 2023, Manuscript No. JAE-23-111384;
Editor assigned date: 29 August, 2023, PreQC No. JAE-23-111384 (PQ);
Reviewed date: 12 September, 2023, QC No. JAE-23-111384;
Revised date: 19 September, 2023, Manuscript No. JAE-23-111384 (R);
Published date: 26 September, 2023, DOI: 10.4172/2324-9080.1000084
Citation: Christopher Collins (2023) DNA Methylation on PPARG and its Relation to Sports Injury and Pain in Male Football (Soccer) Players at all Levels. J Athl Enhanc 12:4
Abstract
Injuries in English football are a cause of great financial burden to clubs and cause distress to players and coaching staff, therefore novel interventions to help prevent injuries should be considered to aid this burden. 287 male players across multiple levels of the English football leagues were tested using saliva for methylation levels across the gene Peroxisome Proliferator-Activated Receptor- Gamma (PPARG), a known inflammation modulation gene. Player injury status and pain scale for each player was analysed at the point of testing to compare against the PPARG methylation status. The average methylation across PPARG on non-injured players was 0.508 and on those injured (n=78) was 0.4488, the difference between player/s indicating a pain score of 10 against the players indicating a pain score of 0 was +0.2437 towards the lower pain result. Therefore, it is theorised that PPARG hypomethylation occurs in response to injury or/and overtraining to aid in the inflammation processes of the body, this study will require further investigation to establish this link. Follow-up analysis on the same players that indicated injury and a high pain score to see if PPARG methylation returns to normative values upon physical improvement is also a logical step for further analysis.
Keywords
Epigenetics; Genetics; Injury; Football; Soccer injuries; Rehabilitation.
Introduction
On average English premier league teams lost 45 million Great Britain Pound (GBP) due to player injury across a season, therefore novel interventions to help prevent injuries should be considered to aid this financial burden on clubs, improve performance of players and maintain the safe playing and training of players [1]. The following research project is completed on 287 male players across multiple levels of the English football leagues from amateur players (193), semiprofessional/professional up to English football league 2 (59), and professional players which are rated from English league 2 and above (35). Injury data was gathered for players regarding injury status and current pain level, and this is correlated against the methylation levels on the gene Peroxisome Proliferator-Activated Receptor-Gamma (PPARG).
Peroxisome Proliferator-Activated Receptor Gamma (PPARG)
The gene Peroxisome Proliferator-Activated Receptor Gamma (PPARG) provides instructions for making a protein that is part of a group of proteins known as nuclear receptors. These receptors are involved in regulating the expression of various genes, which in turn affects different biological processes in the body. The role of the PPARG gene suggests it may impact muscle inflammation and repair processes, especially post musculoskeletal injury due to its ability regulate inflammation. Activation or expression of PPARG is associated with anti-inflammatory processes and the modulation of pro-inflammatory genes [2].
DNA methylation
DNA methylation is a chemical modification of DNA that plays a crucial role in gene regulation and genome stability. It involves the addition of a methyl group (-CH3) to the DNA molecule, specifically at the carbon 5 position of cytosine residues in CpG dinucleotides (where C represents cytosine and G represents guanine). This modification is catalysed by enzymes called DNA methyltransferases [3].
DNA methylation is known as an epigenetic mechanism that can influence gene expression without altering the underlying DNA sequence. Methylation of DNA typically leads to the suppression of gene expression by blocking the binding of transcription factors and other regulatory proteins to the DNA, known as hypermethylation.
DNA methylation patterns are established during early development and are then faithfully maintained throughout cell divisions [4]. Studies have shown DNA methylation patterns can also undergo changes in response to various environmental factors, developmental processes, and diseases [5,6]. DNA methylation patterns have been associated with various diseases, including cancer. In cancer cells, abnormal DNA methylation can lead to the silencing of tumour suppressor genes, allowing uncontrolled cell growth and proliferation [7].
DNA methylation analysis can be performed using various types of biological samples with many studies opting for tissue specific samples, however this incurs disadvantages such as being invasive and lowering the number of recruited participants [8]. One less/non- invasive method to achieve DNA methylation is the usage of saliva. Saliva contains cells from the oral cavity, which can provide DNA for methylation analysis, the sample from saliva is aimed to reproduce systemic DNA methylation.
Methodology
The aim of the study was to analyse if the average methylation across 53 CpG sites on the gene PPARG is indicative of player injury status. Injury status was established with:
1. The player injury records at the time of testing. (Any soft tissue injury that lead to missed games or/and training at the time of testing).
2. A current pain score of 0-10 at the time of testing.
287 anonymised male soccer players (Ages 19-38) were analysed during a period of 21 days over the 2018-2019 English football season (appendix 1); 193 amateur players (unpaid players, but still play in some form of league which includes university/college leagues); 59 semi-professional or professional players (players which are paid on a part time basis or must supplement their football salary with other work); 35 professional players (players where football is their fulltime career).
The study employed the Muhdo Health epigenetic database and tag bio analytics software. Players were tested with saliva and used the Illumina Infinium methylation EPIC V1 array covering ~850k CpG sites, samples were analysed on the iScan high throughput system with the aid of Euro-fins. The average beta value was taken from the raw data for each player on the required CpG sites for PPARG, this measurement on each site is between 0-1, 0 indicating hypo-methylation and 1 being hyper-methylation. The data was ran through the Muhdo Health algorithm tools and players utilised the Muhdo Health mobile phone application to fill out relevant questions.
The 53 CpG sites analysed across PPARG
In Table 1 the pain scale utilised was from 0-10: 0=No pain; 1=No real pain, hardly noticeable; 2=Minor pain on exertion; 3=Minor pain on exertion/may affect daily activities; 4=Minor pain with or without exertion that affects daily activity; 5=Minor pain that rises to moderate pain on exertion; 6=Constant moderate pain; 7=Constant moderate pain that significantly impacts daily activity 8=Constant moderate pain rising to significant pain on exertion; 9=constant significant pain; 10=Unbearable pain/no pain worse.
| CpG sites |
|---|
| cg00057836 |
| cg01412654 |
| cg02430720 |
| cg02605957 |
| cg02726798 |
| cg04702010 |
| cg04908300 |
| cg05563966 |
| cg05671501 |
| cg06340600 |
| cg06573644 |
| cg07424807 |
| cg07556134 |
| cg07676920 |
| cg07895576 |
| cg08288126 |
| cg09405169 |
| cg09663755 |
| cg09702791 |
| cg10237171 |
| cg10441555 |
| cg10499651 |
| cg11176018 |
| cg12880856 |
| cg13369760 |
| cg13518792 |
| cg14113891 |
| cg15722404 |
| cg15938746 |
| cg16197186 |
| cg16340410 |
| cg16470128 |
| cg16827534 |
| cg17369845 |
| cg17819501 |
| cg18063278 |
| cg18303782 |
| cg18861661 |
| cg18887186 |
| cg19046290 |
| cg19182242 |
| cg21859053 |
| cg21946299 |
| cg23514324 |
| cg24271538 |
| cg25845026 |
| cg25911248 |
| cg25929976 |
| cg26197008 |
| cg26364899 |
| cg26741988 |
| cg27051533 |
| cg27095527 |
Table 1: Analysed Cytosine and Guanine (CpG) sites.
Results
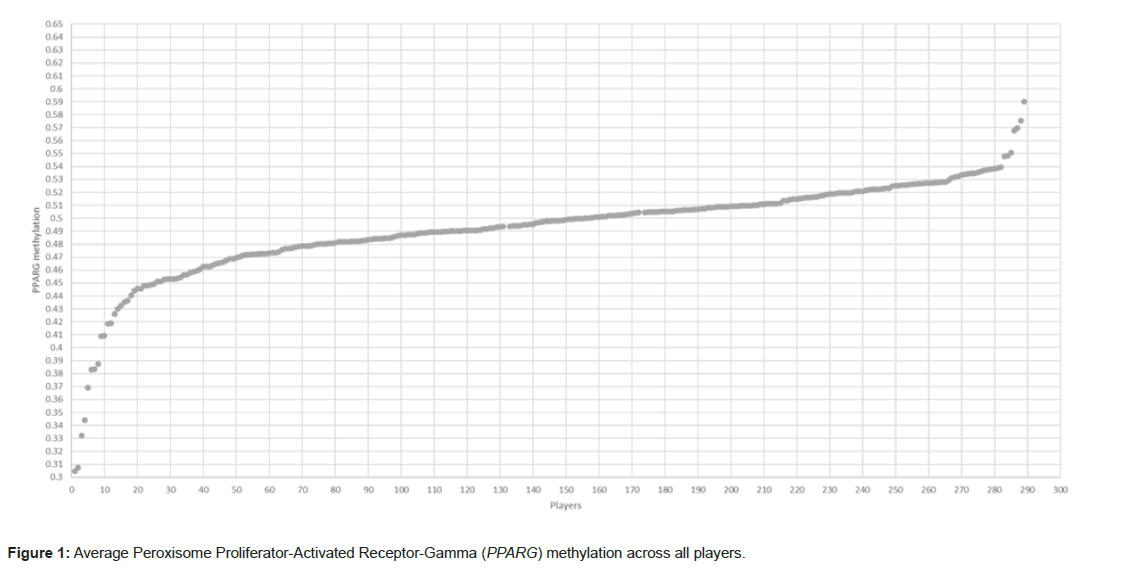
Across all players the average methylation for all CpG sites was 0.4921, Figure 1 shows how most players fall within this range of methylation and this shows some correlation with the Muhdo Health epigenetic database norm based on ~6000 subjects of 0.51 across the same CpG site averages (Figure 1).
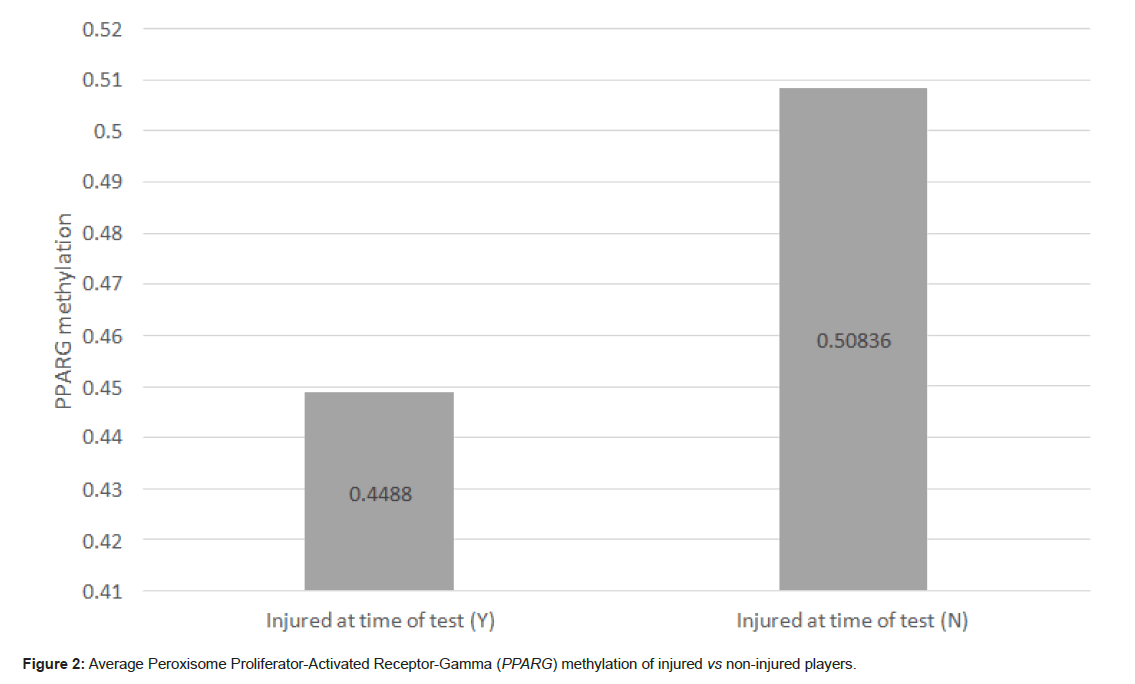
78 players were indicated to have a soft tissue injury that prevented them from play or training at the time of testing, with the remaining 209 indicated to have no injury at time of testing. Figure 2 shows that the average methylation across PPARG on non-injured players was 0.508 and on those injured was 0.4488 (Figure 2).
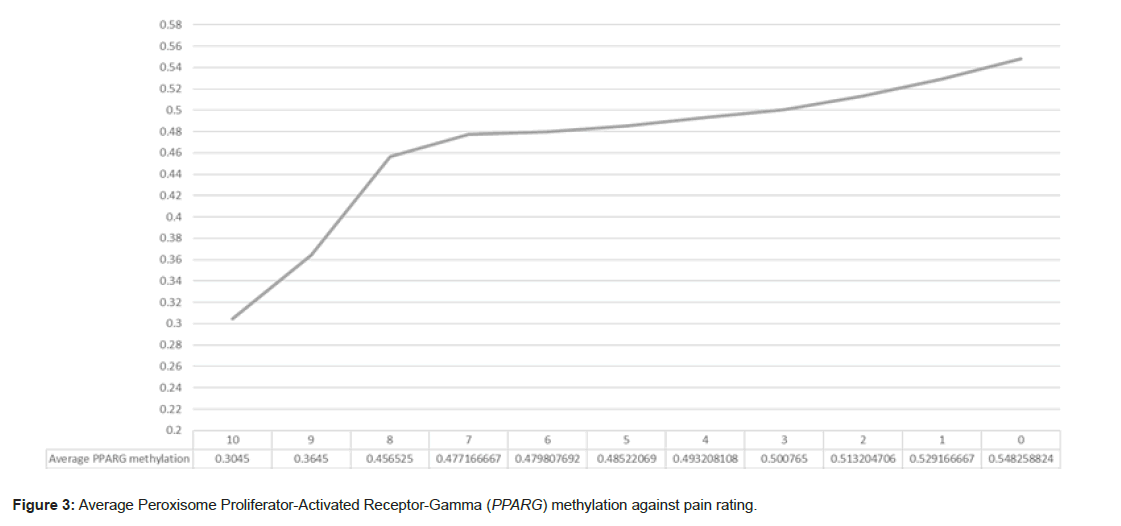
1 player stated they had a pain rating of 10, 8 players stated they had a pain rating of 9, 56 players stated the had a pain rating of 8, 3 players stated they had a pain rating of 7, 13 players stated they had a pain rating of 6, 29 players stated they had a pain rating of 5, 37 players stated they had a pain rating of 4, 20 players stated had a pain rating of 3, 85 players stated they had a pain rating of 2, 18 players stated they had a pain rating of 1 and 17 players stated they had a pain rating of 0. Figure 3 shows that on average those with an indicated pain rating of 5 and greater had a lower PPARG methylation when compared to the overall average PPARG methylation level of 0.4921. The difference between the 1 player indicating a pain score of 10 against the players indicating a pain score of 0 was +0.2437 towards the lower pain result. The average methylation of players indicating a pain score of 0-5 was 0.516920661, the average methylation of players indicating a pain score of 5+ was 0.427953341, a difference of +0.088 towards the average lowest pain scores (Figure 3).
Discussion
The results indicate that lower PPARG methylation is associated with increased indicated pain levels and a higher incidence of injury. Lower levels of methylation are associated with heightened gene expression [9]. As PPARG is associated with anti-inflammatory properties and the modulation of inflammatory markers it would be theorised that lower methylation should be associated with superior pain outcomes and lower injury incidence due to superior expression [10]. However, acute PPARG activation may occur in response to inflammation, injury or illness [11]. PPARG methylation may alter based on a need to modulate pro-inflammation genes, therefore a rapid hypo-methylation may occur in a response to injury or illness, research does show that methylation may alter rapidly [12]. In adipocytes increased methylation across PPARG showed to be the earliest indicator of insulin resistance, whilst tissue specific, this may show acute changeable characteristics of PPARG methylation [13].
This study did not analyse other parameters which may have been affected by PPARG such as diabetes risk, activation of PPARG is known to have blood sugar lowering affects and this may correlate with lower methylation levels across the gene [14]. Compared with the Muhdo Health epigenetic database (~6000) which indicated an average PPARG result of 0.51 the players which indicated the highest pain ratings had a lower PPARG methylation average which backs up a theory that PPARG shows hypo-methylation in response to pain causing pathology to help modulate pro-inflammatory genes. Three players had no indication of injury stopping their play, but their specified pain scale rating was 6 and their methylation level across PPARG was similar to that of players with indicated injury, this increased perceived pain level would in theory result in worsened performance therefore acute hypo-methylation changes in PPARG may be an early warning sign of overtraining or/and low-level injury [15]. One limiting factor to the study could be proposed that the usage of saliva is not as specific as a direct muscle cell methylation pattern, however due to invasiveness this would be impossible given the researcher resources and possible harm to the players. Saliva methylation patterns have shown to be highly correlated to blood methylation patterns and therefore both are legitimate and interchangeable for reproducing this study in other athlete groups [16]. Other variable data was acquired during the study; however, it was not analysed for these results, these variables which include diet, supplements and training schedule may impact PPARG methylation patterns but were not considered during in the analysis.
Follow up from this study aims to analyse player PPARG methylation upon reduction of pain levels or/and change to injury status in players with these indications.
Conclusion
This study shows that there may be a link between PPARG methylation levels and injury status and pain perception in male football players, it is unlikely that PPARG methylation is a causative factor of pain and/or injury due to its inflammatory modulation effects. Therefore, it is theorised that PPARG hypo-methylation occurs in response to injury or/and overtraining to aid in the inflammation processes of the body, this study will require further investigation to establish this link and follow-up analysis on the same players that indicated injury and a high pain score to see if PPARG methylation returns to normative values upon physical improvement. If a link can be adequately established, periodic testing of players for clubs with the financial means to do so may benefit in early prediction of potential injury outcomes and may better monitor players who are falling into overtraining syndrome.
Acknowledgements
Muhdo Health Ltd, for providing access to the epigenetic database and algorithms required for analysis. Tag Bio for analytics software. Euro-fins for providing lab testing and Illumina for providing epigenetic analysis array technology.
References
- Eliakim E, Morgulev E, Lidor R, Meckel Y (2020) Estimation of injury costs: Financial damage of english premier league teams’ underachievement due to injuries. BMJ Open Sport Exerc Med 6(1):e000675.
- Bouhlel MA, Derudas B, Rigamonti E, Dièvart R, Brozek J, et al (2007) PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab 6(2):137-143.
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16(1):6-21.
- Jones PA, Takai D (2001) The role of DNA methylation in mammalian epigenetics. Science 293(5532):1068-1070.
- Suzuki MM, Bird A (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 9(6):465-476.
- Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11(9):607-620.
- Lakshminarasimhan R, Liang G (2016) The role of DNA methylation in cancer. Adv Exp Med Biol 151-172.
- Esteller M (2008) Epigenetics in cancer. N Engl J Med 358(11):1148-1159.
- Counts JL, Goodman JI (1995) Hypomethylation of DNA: a possible epigenetic mechanism involved in tumor promotion. Prog Clin Biol Res 391:81-101.
- Crosby MB, Zhang J, Nowling TM, Svenson JL, Nicol CJ, et al (2006) Inflammatory modulation of PPARγ expression and activity. Clin Immunol 118(2-3):276-283.
- Cai W, Yang T, Liu H, Han L, Zhang K, et al (2018) Peroxisome proliferator-activated receptor γ (PPARγ): A master gatekeeper in CNS injury and repair. Prog Neurobiol 163:27-58.
- Verlinden I, Güiza F, Derese I, Wouters PJ, Joosten K, et al (2020) Time course of altered DNA methylation evoked by critical illness and by early administration of parenteral nutrition in the paediatric ICU. Clin Epigenetics 12:1-0.
- Małodobra-Mazur M, Cierzniak A, Kaliszewski K, Dobosz T (2021) PPARG hypermethylation as the first epigenetic modification in newly onset insulin resistance in human adipocytes. Genes 12(6):889.
- Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S (2011) The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J Adv Pharm Technol Res 2(4):236.
- Kabiri S, Choi J, Shadmanfaat SM, Lee J (2021) Control deficits, conditioning factors, and playing through pain and injury among Iranian professional soccer players. Int J Environ Res Public Health 18(7):3387.
- Langie SA, Moisse M, Declerck K, Koppen G, Godderis L, et al (2017) Salivary DNA methylation profiling: aspects to consider for biomarker identification. Basic Clin Pharmacol Toxicol 121:93-101.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi