Research Article, J Biochem Physiol Vol: 1 Issue: 1
Four CYP19A1 Polymorphisms and Breast Cancer Risk: A MetaAnalysis
Yougen Wu1*, Xiaofeng Qu2, Ju Xia1, Yuting Gu1, Qingqing Qian1,2 and Yang Hong1,3*
1National Institute of Clinical Research, The Fifth People’s Hospital of Shanghai, Fudan University, Shanghai 200240, China
2Department of Pharmacy, The Fifth People’s Hospital of Shanghai, Fudan University, Shanghai 200240, China
3Department of Osteology, The Fifth People’s Hospital of Shanghai, Fudan University, Shanghai 200240, China
*Corresponding Author : Yougen Wu
National Institute of Clinical Research, The Fifth People’s Hospital of Shanghai, Fudan University, Shanghai 200240, China
Tel: +86 21 24289472
Fax: +86 21 24289472
E-mail: wu05055225@126.com
Yang Hong
National Institute of Clinical Research, The Fifth People’s Hospital of Shanghai, Fudan University, Shanghai 200240, China
Tel: +86 21 24289472
Fax: +86 21 24289472
E-mail: hongyang@fudan.edu.cn
Received: February 07, 2018 Accepted: February 22, 2018 Published: February 28, 2018
Citation: Wu Y, Qu X, Xia J, Gu Y, Qian Q, et al. (2018) Four CYP19A1 Polymorphisms and Breast Cancer Risk: A Meta-Analysis. J Biochem Physiol 1:1.
Abstract
Many molecular epidemiological studies have investigated an association between CYP19A1 gene single-nucleotide polymorphisms (SNPs) and breast cancer risk, but results have remained controversial and inconclusive. In order to reveal the real association, we performed an updated meta-analysis including two CYP19A1 gene polymorphisms (rs700519, rs10046). Moreover, we performed a meta-analysis of another two CYP19A1 (rs2236722 and rs4646) gene polymorphisms for the first time to evaluate their relevance in susceptibility to breast cancer risk. A systematic database search was conducted to retrieve eligible articles. The odds ratio (OR) with 95% confidence interval (95% CI) were used to assess the strength of the association.A total of 38 eligible studies were included in the meta-analysis, and the results showed that three CYP19A1 gene polymorphisms (rs700519, rs10046, and rs2236722) had no relationship with an increased/decreased breast cancer risk in overall or ethnicity-based populations (all P values were more than 0.05); CYP19A1 rs4646 polymorphism was significant associated with an increased breast
cancer risk in overall populations under dominant genetic model (CC+AC vs. AA, OR=1.179, 95% CI=1.056 - 1.315, P-value=0.003). However, we did not find an association between CYP19A1 rs4646 polymorphism and breast cancer susceptibility among Asian populations (P value was more than 0.05).The meta-analysis indicates that CYP19A1 rs4646 polymorphism may be associated with breast cancer risk. Further epidemiological studies with larger sample sizes are needed to validate the association between CYP19A1 rs4646 polymorphism and breast cancer risk in various populations.
Keywords: CYP19A1; Polymorphism; Breast cancer; Risk; Meta-analysis
Introduction
Breast cancer is the most common malignancy among women worldwide. Numerous studies suggest that breast carcinogenesis and progression is influenced by steroid hormones, particularly estrogens [1,2].
Some genetic variations of steroid hormone pathway genes involved in the metabolism of androgens and estrogens are associated with the risk of breast cancer [3,4]. The cytochrome P450 family 19 subfamily a member 1 (CYP19A1) gene is located on chromosome 15q21.2 region and encodes aromatase, which converts androstenedione and testosterone into estrone and estradiol, respectively [5]. CYP19A1 mutations can alter aromatase activity, which affects estrogen levels indirectly, and may ultimately alter susceptibility to breast cancer [6,7].
To date, an increasing number of studies have evaluated the potential association between the CYP19A1 polymorphisms and the risk of breast cancer in diverse populations. Four CYP19A1 genetic polymorphisms including rs700519 (Arg264Cys) located in exon 7 codon 264, the rs10046 located in the 3’ untranslated region (3’- UTR), CYP19A1 polymorphism at codon 39 Trp/Arg (rs2236722), and the rs4646 located in the 3’-UTR have been focused on a large scale. However, the results are inconsistent and inconclusive.
One previous meta-analysis suggested no association between CYP19A1 rs700519 polymorphism and breast cancer risk [6], and another meta-analysis indicated that rs10046 polymorphism on CYP19A1 did not affect breast cancer risk [8]. However, limited studies were included in both meta-analyses. Recently, several more studies assessing the association between the CYP19A1 polymorphisms (rs700519 and rs10046) and breast cancer risk have been published. We therefore conducted an updated meta-analysis to clarify the association of the CYP19A1 polymorphisms (rs700519 and rs10046) with risk of breast cancer in different populations. In addition, we performed a meta-analysis of another two CYP19A1 (rs2236722 and rs4646) gene polymorphisms for the first time to evaluate their relevance in susceptibility to breast cancer risk.
Materials and Methods
Literature and search strategy
PubMed, Web of Science and Embase database were searched (until April 30, 2017) for eligible articles. The search strategy used combinations of the following keywords: “CYP19” or “CYP19A1” and “polymorphism” or “variant” or “mutation” and “breast cancer”.
Inclusion and exclusion criteria
Eligible studies had to meet the following criteria: (1) studies addressed the potential association of four CYP19A1 genetic polymorphisms [rs700519, rs10046, rs2236722, and rs4646] and breast cancer risk, (2) studies based on case–control design and (3) studies with sufficient data about genotype distribution of controls and cases. The exclusion criteria were: (1) studies with no sufficient data about genotype distribution of controls and cases, (2) duplicate publications and (3) comments, case reports, abstract and review articles (including meta-analysis).
Data extraction
The following data was extracted: (1) name of the first author, (2) year of publication, (3) country of origin, (4) ethnicity, (5) source of control groups ( hospital-based or population-based controls or mixed), (6) number of genotyped cases and controls, Data was extracted from the final selected studies independently by two authors.
Statistical analysis
The relationship between CYP19A1 polymorphisms and breast cancer risk was assessed by a combined odds ratio (OR) with corresponding 95% confidence interval (95% CI) under co-dominant model, dominant model, and recessive model, respectively. Subgroup analyses based on ethnicity (Caucasians/Asians) was performed. The significance of the pooled OR estimate was determined by a Z test. The statistical significance was set at p value < 0.05.
Cochran’s chi-square-based Q and I2 statistics were used to evaluate heterogeneity across studies. If heterogeneity did not exist (P value > 0.1 for the Q test) among studies, the fixed effects model was used [9]; otherwise, the random effects model was applied [10]. I2 statistic was calculated to quantify the proportion of the total heterogeneity among studies. Generally, I2 values of 75%, 50%, and 25% indicated high, moderate, and low heterogeneity, respectively.
Sensitivity analysis was conducted to assess the influence of each study on the overall estimate by excluding studies one by one and recalculating the combined results of the remaining studies.
Publication bias of literatures was detected by funnel-plot analysis and Egger’s test [11]. Data analyzes were performed with STATA version 11.0 (Stata Corporation, College Station, Texas, USA).
Results
Characteristics of the studies
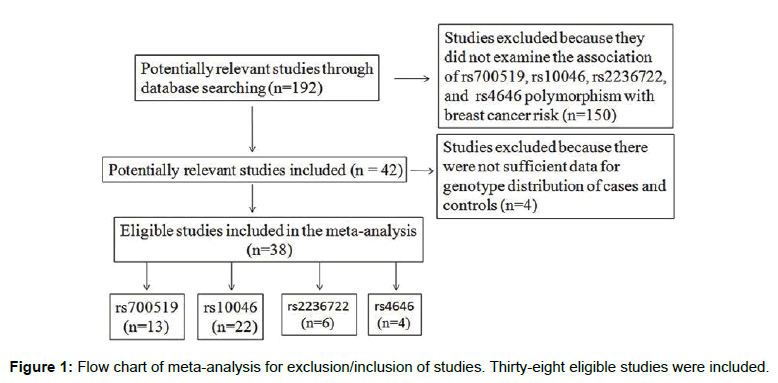
We retrieved a total of 38 studies according to the inclusion/ exclusion criteria, of which included a total of 13 studies containing 4,099 cases and 5,624 controls for the rs700519 polymorphism (Table 1) [12-24] , 22 studies containing 12,589 cases and 17,277 controls referring to the rs10046 polymorphism (Table 2) [7,8,23-39], 6 studies with 957 cases and 1,368 controls involved in the rs2236722 polymorphisms (Table 3) [12,40-44], and 4 studies with 4,970 cases and 5,925 controls involved in the rs4646 polymorphism (Table 4) [28,38,45,46]. A detailed flow chart of the exclusion and inclusion process was showed in Figure 1.
| First author (Year) | Country | Ethnicity | Source | Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CT+TT | CC+CT | CC | CT | TT | CT+TT | CC+CT | ||||
| Miyoshi (2000) [12] | Japan | Asian | H | 109 | - | - | 89 | - | 85 | - | - | 93 | - |
| Lee (2003) [13] | Korea | Asian | H | 150 | 134 | 4 | 138 | 284 | 176 | 106 | 6 | 112 | 282 |
| Hefler (2004) [14] | Austria | Caucasian | P | 367 | 22 | 0 | 22 | 389 | 1503 | 107 | 9 | 116 | 1610 |
| Song (2006) [15] | China | Asian | P | 84 | 22 | 2 | 24 | 106 | 87 | 24 | 1 | 25 | 111 |
| Hu (2007) [16] | China | Asian | H | 87 | 24 | 1 | 25 | 111 | 84 | 22 | 2 | 24 | 106 |
| Gulyaeva (2008) [17] | Russia | Caucasian | H | 100 | 8 | 0 | 8 | 108 | 168 | 10 | 4 | 14 | 178 |
| Justenhoven (2008) [18] | Germany | Caucasian | P | 549 | 49 | 1 | 50 | 598 | 561 | 60 | 1 | 61 | 621 |
| Sangrajrang (2009) [19] | Thailand | Asian | H | 331 | 201 | 31 | 232 | 532 | 297 | 167 | 19 | 186 | 464 |
| Wang (2009) [20] | China | Asian | H | 97 | 78 | 25 | 103 | 175 | 98 | 77 | 25 | 102 | 175 |
| Khvostova (2012) [21] | Russia | Caucasian | H | 283 | 39 | 1 | 40 | 322 | 477 | 57 | 2 | 59 | 534 |
| Chattopadhyay (2014) [22] | India | Asian | P | 226 | 115 | 19 | 134 | 341 | 258 | 91 | 11 | 102 | 349 |
| Sun (2015) [23] | China | Asian | H | 410 | 111 | 9 | 120 | 521 | 392 | 143 | 11 | 154 | 535 |
| Pan (2016) [24] | China | Asian | H | 225 | 87 | 9 | 96 | 312 | 289 | 96 | 5 | 101 | 385 |
Table 1: Characteristics of case–control studies included in CYP19A1 R264C polymorphism (rs700519) and breast cancer risk.
| First author (Year) | Country | Ethnicity | Source | Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CT+TT | CC+CT | CC | CT | TT | CT+TT | CC+CT | ||||
| Kristensen (2000) [25] | Norway | Caucasian | HP | 95 | 240 | 146 | 386 | 335 | 69 | 114 | 53 | 167 | 183 |
| Haiman (2002) [26] | US | Caucasian | H | 103 | 240 | 118 | 358 | 343 | 134 | 310 | 167 | 477 | 444 |
| Dunning (2004) [7] | UK | Caucasian | H | 610 | 1286 | 739 | 2025 | 1896 | 808 | 1773 | 1049 | 2822 | 2581 |
| Ralph-1 (2007) [27] | US | Caucasian | H | 349 | 830 | 461 | 1291 | 1179 | 758 | 1650 | 883 | 2533 | 2408 |
| Ralph-2 (2007) [27] | US | Caucasian | H | 129 | 231 | 142 | 373 | 360 | 222 | 503 | 274 | 777 | 725 |
| Chen (2008) [28] | China | Asian | H | 125 | 308 | 178 | 486 | 433 | 163 | 436 | 277 | 713 | 599 |
| Zhang (2008) [29] | China | Asian | H | 55 | 151 | 94 | 245 | 206 | 94 | 176 | 120 | 296 | 270 |
| Iwasaki-1 (2009) [30] | Japan | Asian | H | 118 | 188 | 82 | 270 | 306 | 125 | 194 | 69 | 263 | 319 |
| Iwasaki-2 (2009) [30] | Japan | Asian | H | 24 | 41 | 14 | 55 | 65 | 22 | 44 | 13 | 57 | 66 |
| Iwasaki-3 (2009) [30] | Brasil | Caucasian | H | 133 | 179 | 67 | 246 | 312 | 121 | 200 | 58 | 258 | 321 |
| Yoshimoto (2011) [31] | Japan | Asian | H | 239 | 427 | 160 | 587 | 666 | 97 | 120 | 60 | 180 | 217 |
| Pineda (2012) [8] | Spain | Caucasian | H | 135 | 278 | 109 | 387 | 413 | 281 | 629 | 311 | 940 | 910 |
| Clendenen (2013) [32] | US and Sweden | Mixed | P | 306 | 548 | 308 | 856 | 854 | 549 | 1032 | 523 | 1555 | 1581 |
| Iwasaki (2013) [33] | Japan | Asian | H | 116 | 253 | 253 | - | 117 | 252 | 252 | - | ||
| Ghisari (2014) [34] | Denmark | Caucasian | P | 23 | 8 | 0 | 8 | 31 | 79 | 29 | 6 | 35 | 108 |
| Zins (2014) [35] | Austria | Caucasian | P | 65 | 142 | 67 | 209 | 207 | 55 | 136 | 62 | 198 | 191 |
| Sun (2015) [23] | China | Asian | H | 111 | 264 | 155 | 419 | 375 | 126 | 290 | 130 | 420 | 416 |
| Yang (2015) [36] | China | Asian | H | 30 | 48 | 34 | 82 | 78 | 25 | 82 | 32 | 114 | 107 |
| Pan (2016) [24] | China | Asian | H | 49 | 185 | 100 | 285 | 234 | 89 | 192 | 111 | 303 | 281 |
| Farzaneh (2016) [37] | Iran | Asian | H | 23 | 68 | 33 | 101 | 91 | 30 | 55 | 15 | 70 | 85 |
| Kopp (2016) [38] | Denmark | Caucasian | P | 159 | 346 | 182 | 528 | 505 | 146 | 353 | 188 | 541 | 499 |
| Ghisari (2017) [39] | Denmark | Caucasian | P | 36 | 68 | 38 | 106 | 104 | 47 | 93 | 56 | 149 | 140 |
Table 2: Characteristics of case–control studies included in CYP19A1 polymorphism (rs10046) and breast cancer risk.
| First author (Year) | Country | Ethnicity | Source | Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | CT+CC | TT+CT | TT | CT | CC | CT+CC | TT+CT | ||||
| Miyoshi (2000) [12] | Japan | Asian | H | 195 | - | - | 8 | - | 180 | - | - | 19 | - |
| Hirose (2004) [40] | Japan | Asian | H | 227 | 20 | 1 | 21 | 247 | 561 | 38 | 4 | 42 | 599 |
| Sobczuk (2009) [41] | Poland | Caucasian | H | 20 | 45 | 35 | 80 | 65 | 18 | 58 | 30 | 88 | 76 |
| TÜZÜNER (2010) [42] | Turkey | Caucasian | P | 3 | 52 | 0 | 52 | 55 | 27 | 64 | 0 | 64 | 91 |
| Ramalhinho (2012) [43] | Portugal | Caucasian | H | 40 | - | - | 61 | - | 65 | - | - | 56 | - |
| Surekha (2014) [44] | India | Asian | P | 227 | 23 | 0 | 23 | 250 | 170 | 78 | 0 | 78 | 248 |
Table 3: Characteristics of case–control studies included in CYP19A1 polymorphism (rs2236722) and breast cancer risk.
| First author (Year) | Country | Ethnicity | Source | Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | AC | AA | AC+AA | CC+AC | CC | AC | AA | AC+AA | CC+AC | ||||
| Chen (2008) [28] | China | Asian | H | 298 | 260 | 53 | 313 | 558 | 441 | 358 | 77 | 435 | 799 |
| Boone (2014) [45] | US | Caucasian | P | - | - | 540 | - | 2984 | - | - | 756 | - | 3452 |
| Alanazi (2015) [46] | Kingdom of Saudi Arabia | Asian | P | 94 | 46 | 8 | 54 | 140 | 99 | 47 | 8 | 55 | 146 |
| Kopp (2016) [38] | Denmark | Caucasian | P | 372 | 265 | 50 | 315 | 637 | 371 | 262 | 54 | 316 | 633 |
Table 4: Characteristics of case–control studies included in CYP19A1 polymorphism (rs4646) and breast cancer risk.
Quantitative synthesis
The summary of meta-analysis and heterogeneity test results for CYP19A1 polymorphisms with breast cancer risk were presented in Table 5. For rs700519 polymorphism, no significant associations were found with the risk of breast cancer in overall or race-based populations in any of the genetic models tested. For CYP19A1 rs10046 polymorphism, we found no significant association with breast cancer risk in overall population. The analysis by racial/ethnic subgroups also failed to produce significant associations in any of the genetic models tested. Furthermore, we observed no significant association for CYP19A1 rs2236722 polymorphism with the risk of breast cancer in overall or ethnicity-based populations. I2 > 75.0 % was observed in overall analyses. Sensitivity analysis was conducted to investigate the influence of each study on the overall pooled OR. The exclusion of Surekha et al., 2014 study made the biggest drop for heterogeneity values and still no significant association of the CYP19A1 rs2236722 polymorphism with breast cancer risk was observed (data not shown).
| Polymorphisms | Comparisons | No. of studies | Sample size | OR [95% CI] | P value | I2 (P) | Model | |
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||
| CYP19A1 R264C | ||||||||
| Overall | TT vs. CC | 12 | 3011 | 4486 | 1.178 [0.877, 1.583] | p=0.277 | 0.0% (p=0.638) | F |
| TC vs. CC | 12 | 3799 | 5350 | 1.062 [0.951, 1.185] | p=0.286 | 33.1% (p=0.125) | F | |
| TT+TC vs. CC | 13 | 4099 | 5624 | 1.034 [0.894, 1.196] | p=0.653 | 43.9% (p=0.045) | R | |
| TT vs. TC+CC | 12 | 3901 | 5446 | 1.141 [0.853, 1.526] | p=0.374 | 0.0% (p=0.696) | F | |
| Caucasian | TT vs. CC | 4 | 1301 | 2725 | 0.387 [0.107, 1.398] | p=0.148 | 0.0% (p=0.735) | F |
| TC vs. CC | 4 | 1417 | 2943 | 0.950 [0.747, 1.207] | p=0.672 | 0.0% (p=0.586) | F | |
| TT+TC vs. CC | 4 | 1419 | 2959 | 0.909 [0.718, 1.152] | p=0.431 | 0.0% (p=0.634) | F | |
| TT vs. TC+CC | 4 | 1419 | 2959 | 0.386 [0.107, 1.394] | p=0.146 | 0.0% (p=0.733) | F | |
| Asian | TT vs. CC | 8 | 1710 | 1761 | 1.283 [0.942, 1.747] | p=0.114 | 0.0% (p=0.588) | F |
| TC vs. CC | 8 | 2382 | 2407 | 1.102 [0.920, 1.322] | p=0.292 | 48.2% (p=0.061) | R | |
| TT+TC vs. CC | 9 | 2680 | 2665 | 1.073 [0.895, 1.287] | p=0.447 | 55.9% (p=0.020) | R | |
| TT vs. TC+CC | 8 | 2482 | 2487 | 1.236 [0.913, 1.674] | p=0.171 | 0.0% (p=0.650) | F | |
| rs10046 | ||||||||
| Overall | TT vs. CC | 21 | 6144 | 8497 | 1.058 [0.951, 1.177] | p=0.297 | 46.8% (p=0.010) | R |
| TC vs. CC | 21 | 8993 | 12451 | 1.020 [0.932, 1.117] | p=0.668 | 45.2% (p=0.013) | R | |
| TT+TC vs. CC | 22 | 12589 | 17277 | 1.030 [0.946, 1.121] | p=0.498 | 46.9% (p=0.008) | R | |
| TT vs. TC+CC | 21 | 12220 | 16908 | 1.022 [0.969, 1.079] | p=0.423 | 25.6% (p=0.139) | F | |
| Caucasian | TT vs. CC | 11 | 4735 | 6710 | 0.979 [0.855, 1.120] | p=0.755 | 47.6% (p=0.039) | R |
| TC vs. CC | 11 | 5685 | 8510 | 0.975 [0.907, 1.050] | p=0.506 | 16.4% (p=0.288) | F | |
| TT+TC vs. CC | 11 | 7754 | 11617 | 0.969 [0.876, 1.071] | p=0.533 | 38.4% (p=0.093) | R | |
| TT vs. TC+CC | 11 | 7754 | 11617 | 0.993 [0.930, 1.061] | p=0.838 | 22.3% (p=0.231) | F | |
| Asian | TT vs. CC | 9 | 1624 | 1598 | 1.219 [0.997, 1.490] | p=0.053 | 41.4% (p=0.091) | R |
| TC vs. CC | 9 | 2454 | 2360 | 1.139 [0.924, 1.404] | p=0.224 | 59.4% (p=0.011) | R | |
| TT+TC vs. CC | 10 | 3673 | 3556 | 1.145 [0.971, 1.352] | p=0.108 | 50.3% (p=0.034) | R | |
| TT vs. TC+CC | 9 | 3303 | 3187 | 1.081 [0.964, 1.212] | p=0.184 | 32.4% (p=0.158) | F | |
| rs2236722 | ||||||||
| Overall | CC vs. TT | 2 | 283 | 613 | 0.979 [0.464, 2.066] | p=0.956 | 0.0% (p=0.656) | F |
| CT vs. TT | 4 | 617 | 1014 | 1.007 [0.295, 3.436] | p=0.991 | 92.1% (p=0.000) | R | |
| CC+CT vs. TT | 6 | 957 | 1368 | 0.955 [0.404, 2.258] | p=0.916 | 90.1% (p=0.000) | R | |
| CC+CT vs. TTa | 5 | 707 | 1120 | 1.272 [0.644, 2.513] | p=0.489 | 77.8% (p=0.001) | R | |
| CC vs. CT+TT | 2 | 348 | 709 | 1.281 [0.729, 2.251] | p=0.389 | 0.0% (p=0.484) | F | |
| Caucasian | CC vs. TT | 1 | 228 | 565 | 1.050 [0.471, 2.342] | p=0.905 | NA | R |
| CT vs. TT | 2 | 120 | 167 | 2.143 [0.205, 22.448] | p=0.525 | 90.4% (p=0.001) | R | |
| CC+CT vs. TT | 3 | 256 | 318 | 1.931 [0.728, 5.125] | p=0.186 | 78.9% (p=0.000) | R | |
| CC vs. CT+TT | 1 | 100 | 106 | 1.364 [0.757, 2.459] | p=0.302 | NA | R | |
| Asian | CC vs. TT | 1 | 55 | 48 | 0.618 [0.069, 5.558] | p=0.667 | NA | R |
| CT vs. TT | 2 | 497 | 847 | 0.534 [0.094, 3.043] | p=0.479 | 95.3% (p=0.000) | R | |
| CC+CT vs. TT | 3 | 701 | 1050 | 0.475 [0.151, 1.494] | p=0.203 | 90.4% (p=0.000) | R | |
| CC+CT vs. TTa | 2 | 451 | 802 | 0.727 [0.234, 2.254] | p=0.581 | 80.2% (p=0.025) | R | |
| CC vs. CT+TT | 1 | 248 | 603 | 0.606 [0.067, 5.452] | p=0.655 | NA | R | |
| rs4646 | ||||||||
| Overall | CC vs. AA | 3 | 875 | 1050 | 1.022 [0.781, 1.337] | p=0.877 | 0.0% (p=0.933) | F |
| AC vs. AA | 3 | 682 | 806 | 1.065 [0.810, 1.402] | p=0.651 | 0.0% (p=0.980) | F | |
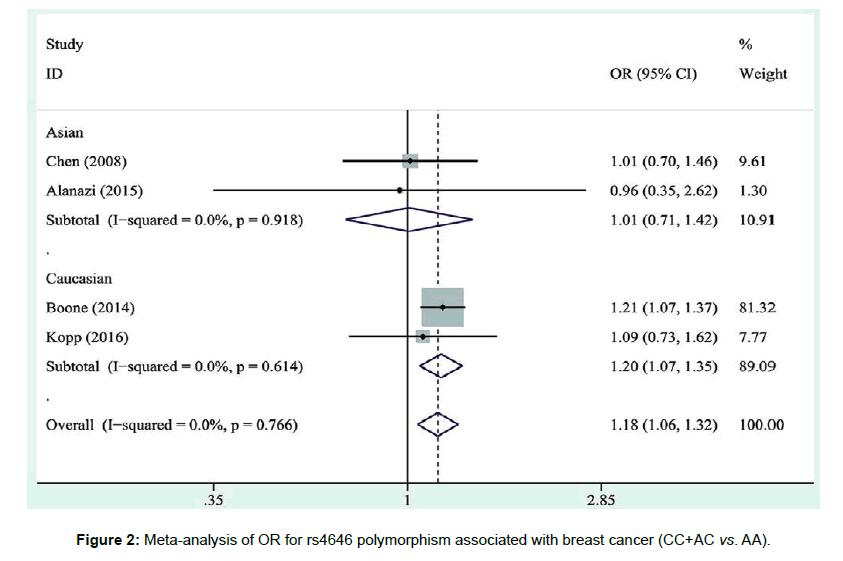
| CC+AC vs. AA | 4 | 4970 | 5925 | 1.179 [1.056, 1.315] | p=0.003 | 0.0% (p=0.766) | F | |
| CC vs. AC+AA | 3 | 1446 | 1717 | 0.971 [0.843, 1.118] | p=0.680 | 0.0% (p=0.902) | F | |
| Caucasian | CC vs. AA | 1 | 422 | 425 | 1.083 [0.718, 1.633] | p=0.704 | NA | R |
| AC vs. AA | 1 | 315 | 316 | 1.092 [0.717, 1.664] | p=0.681 | NA | R | |
| CC+AC vs. AA | 2 | 4211 | 4895 | 1.199 [1.068, 1.346] | p=0.002 | 0.0% (p=0.614) | F | |
| CC vs. AC+AA | 1 | 687 | 687 | 1.006 [0.814, 1.244] | p=0.957 | NA | R | |
| Asian | CC vs. AA | 2 | 453 | 625 | 0.978 [0.685, 1.395] | p=0.901 | 0.0% (p=0.952) | F |
| AC vs. AA | 2 | 367 | 490 | 1.046 [0.729, 1.501] | p=0.807 | 0.0% (p=0.896) | F | |
| CC+AC vs. AA | 2 | 759 | 1030 | 1.008 [0.715, 1.422] | p=0.964 | 0.0% (p=0.918) | F | |
| CC vs. AC+AA | 2 | 759 | 1030 | 0.944 [0.781, 1.140] | p=0.547 | 0.0% (p=0.911) | F | |
F=fixed effects model; R=random effects model.
Table 5: Meta-analysis of CYP19A1 genes polymorphisms and breast cancer risk.
For CYP19A1 rs4646 polymorphism, the meta-analysis showed that individuals with the CC/AC genotype were significantly associated with an increased breast cancer risk as compared with AA genotype in overall or Caucasian populations (Overall: OR=1.179, 95% CI=1.056–1.315; Caucasian: OR=1.199, 95% CI=1.068-1.346). However, we found no evidence of association between CYP19A1 rs4646 polymorphism and susceptibility to breast cancer among Asian women (Table 5, Figure 2).
Potential publication bias
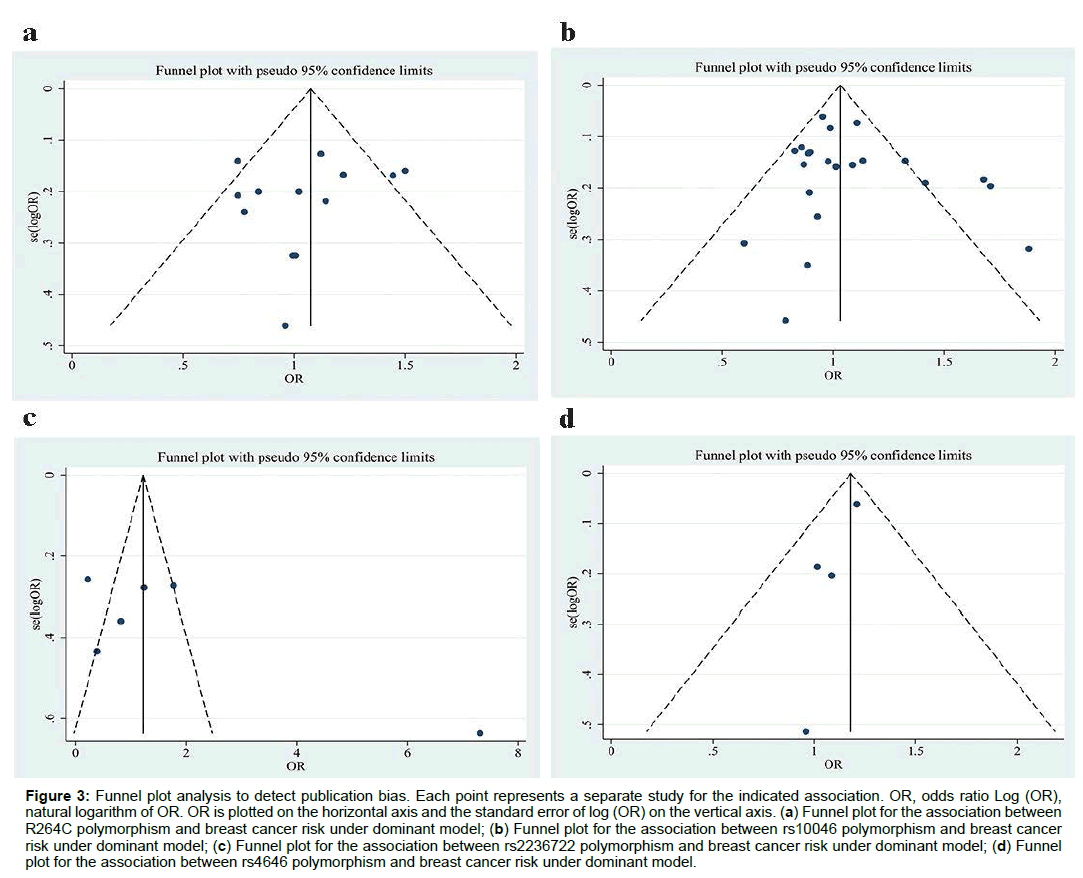
The shape of funnel plot did not show obvious asymmetry (Figure 3). Egger’s test revealed no statistical evidence for publication bias (All P>0.05).
Figure 3: Funnel plot analysis to detect publication bias. Each point represents a separate study for the indicated association. OR, odds ratio Log (OR), natural logarithm of OR. OR is plotted on the horizontal axis and the standard error of log (OR) on the vertical axis. (a) Funnel plot for the association between R264C polymorphism and breast cancer risk under dominant model; (b) Funnel plot for the association between rs10046 polymorphism and breast cancer risk under dominant model; (c) Funnel plot for the association between rs2236722 polymorphism and breast cancer risk under dominant model; (d) Funnel plot for the association between rs4646 polymorphism and breast cancer risk under dominant model.
Discussion
CYP19A1 is a key estrogen biosynthesis enzyme and play an important role in the development of breast cancer. In the current study, we have analyzed an almost 1.63 and 1.83 fold larger number of studies than Ma [6] and Pineda [8], respectively. We found no statistically significant association between breast cancer risk and CYP19A1 polymorphisms (rs700519 and rs10046), which is consistent with the results of the previous meta-analysis for breast cancer [6,8]. Our results confirmed and established the trend of association between the CYP19A1 polymorphisms (rs700519 and rs10046) and breast cancer risk indicated by the meta-analysis of Ma and Pineda [6,8]. To explain the result, we can speculate that the effect of CYP19A1 rs700519 polymorphism on breast cancer risk is limited. CYP19A1 rs700519 polymorphism is not the only factor that influences aromatase activity for estrogens biosynthesis. In fact, R264C and R264H polymorphisms differentially influenced human aromatase activity and function [47].
The present meta-analysis is the first to evaluate the association between CYP19A1 polymorphisms (rs2236722 and rs4646) and breast cancer risk. Pooled analysis found no evidence of association between CYP19A1 polymorphism (rs2236722) and susceptibility to breast cancer. In addition, the sensitivity analysis results showed that Surekha et al., 2014 study was the source of heterogeneity [44]. The conclusion remained unchanged even after the fore-mentioned study was excluded. Overall, the CYP19A1 rs2236722 is a rare polymorphism, the result should be interpreted cautiously owing to the relatively small sample size within these two ethnic populations for CYP19A1 rs2236722 polymorphism. Relationship between CYP19A1 rs2236722 polymorphism and CYP19A1 enzyme activity are also needed for confirmation in the future studies.
It is particularly worth noting that the association of CYP19A1 rs4646 polymorphism with breast cancer risk was observed in overall and Caucasian populations, but not in Asian populations. One possibility is that the sample size for rs4646 among Asian populations is too small to show significant evidence. It is also possible that the effect strength of genetic alterations predisposing to human diseases is different in different racial populations [48].
Conclusion
The present meta-analysis suggests that three variants (rs700519, rs10046, and rs2236722) in the CYP19A1 gene are not significantly associated with breast cancer risk. One SNP (rs4646) may contribute to increasing susceptibility to breast cancer. More well-designed association studies with larger sample size of different ethnic populations will be needed to confirm the risk identified in the current meta-analysis.
Acknowledgement
Financial support: No financial support was received for the study.
Conflict of interest
Yougen Wu and Xiaofeng Qu have contributed equally to the work
The authors declare no conflict of interest.
References
- Saha RS, Vadlamudi RK (2012) Role of estrogen receptor signaling in breast cancer metastasis. Int J Breast Cancer. 2012: 654698.
- Platet N, Cathiard AM, Gleizes M, Garcia M (2004) Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol 51: 55-67.
- Beckmann L, Husing A, Setiawan VW, Amiano P, Clavel-Chapelon F, et al. (2011) Comprehensive analysis of hormone and genetic variation in 36 genes related to steroid hormone metabolism in pre- and postmenopausal women from the breast and prostate cancer cohort consortium (BPC3). J Clin Endocrinol Metab 96: E360-367.
- Nyante SJ, Gammon MD, Kaufman JS, Bensen JT, Lin DY, et al. (2015) Genetic variation in estrogen and progesterone pathway genes and breast cancer risk: an exploration of tumor subtype-specific effects. Cancer Causes Control 26: 121-131.
- Latil AG, Azzouzi R, Cancel GS, Guillaume EC, Cochan-Priollet B, et al. (2001) Prostate carcinoma risk and allelic variants of genes involved in androgen biosynthesis and metabolism pathways. Cancer 92: 1130-1137.
- Ma X, Qi X, Chen C, Lin H, Xiong H, et al. (2010) Association between CYP19 polymorphisms and breast cancer risk: results from 10,592 cases and 11,720 controls. Breast Cancer Res Treat 122: 495-501.
- Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, et al. (2004) Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst 96: 936-945.
- Pineda B, Garcia-Perez MA, Cano A, Lluch A, Eroles P (2013) Associations between aromatase CYP19 rs10046 polymorphism and breast cancer risk: from a case-control to a meta-analysis of 20,098 subjects. PLoS One 8: e53902.
- Dersimonian R, Laird N (2015) Meta-analysis in clinical trials revisited. Contemp Clin Trials 45: 139-145.
- Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719-748.
- Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629-634.
- Miyoshi Y, Iwao K, Ikeda N, Egawa C, Noguchi S (2000) Breast cancer risk associated with polymorphism in CYP19 in Japanese women. Int J Cancer 89: 325-328.
- Lee KM, Abel J, Ko Y, Harth V, Park WY, et al. (2003) Genetic polymorphisms of cytochrome P450 19 and 1B1, alcohol use, and breast cancer risk in Korean women. Br J Cancer 88: 675-678.
- Hefler LA, Tempfer CB, Grimm C, Lebrecht A, Ulbrich E, et al. (2004) Estrogen-metabolizing gene polymorphisms in the assessment of breast carcinoma risk and fibroadenoma risk in Caucasian women. Cancer 101: 264-269.
- Song CG, Hu Z, Yuan WT, Di GH, Shen ZZ, et al. (2006) Effect of R264C polymorphism in CYP19A1 gene on BRCA1/2-negative hereditary breast cancer from Shanghai population of China. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 23: 181-183.
- Hu Z, Song CG, Lu JS, Luo JM, Shen ZZ, et al. (2007) A multigenic study on breast cancer risk associated with genetic polymorphisms of ER Alpha, COMT and CYP19 gene in BRCA1/BRCA2 negative Shanghai women with early onset breast cancer or affected relatives. J Cancer Res Clin Oncol 133: 969-978.
- Gulyaeva LF, Mikhailova ON, Pustyinyak VO, Kim IVT, Gerasimov AV, et al. (2008) Comparative analysis of SNP in estrogen-metabolizing enzymes for ovarian, endometrial, and breast cancers in Novosibirsk, Russia. Adv Exp Med Biol 617: 359-366.
- Justenhoven C, Hamann U, Schubert F, Zapatka M, Pierl CB, et al. (2008) Breast cancer: a candidate gene approach across the estrogen metabolic pathway. Breast Cancer Res Treat 108: 137-149.
- Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Laird NM, et al. (2009) Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. Int J Cancer 125: 837-843.
- Wang YP, Li H, Li JY, Yuan P, Yang F, et al. (2009) Relationship between estrogen-biosynthesis gene (CYP17, CYP19, HSD17beta1) polymorphisms and breast cancer. Zhonghua Zhong Liu Za Zhi 31: 899-903.
- Khvostova EP, Pustylnyak VO, Gulyaeva LF (2012) Genetic polymorphism of estrogen metabolizing enzymes in Siberian women with breast cancer. Genet Test Mol Biomarkers 16: 167-173.
- Chattopadhyay S, Siddiqui S, Akhtar MS, Najm MZ, Deo SV, et al. (2014) Genetic polymorphisms of ESR1, ESR2, CYP17A1, and CYP19A1 and the risk of breast cancer: a case control study from North India. Tumour Biol 35: 4517-4527.
- Sun MY, Du HY, Zhu AN, Liang HY, De Garibay GR, et al. (2015) Genetic polymorphisms in estrogen-related genes and the risk of breast cancer among Han Chinese women. Int J Mol Sci 16: 4121-4135.
- Pan Z, Fu Z, Song Q, Cao W, Cheng W, Xu X (2016) Genetic polymorphisms and haplotype of hormone-related genes are associated with the risk of breast cancer in Chinese women. Genet Mol Res 15.
- Kristensen VN, Harada N, Yoshimura N, Haraldsen E, Lonning PE, et al. (2000) Genetic variants of CYP19 (aromatase) and breast cancer risk. Oncogene 19: 1329-1333.
- Haiman CA, Hankinson SE, Spiegelman D, Brown M, Hunter DJ (2002) No association between a single nucleotide polymorphism in CYP19 and breast cancer risk. Cancer Epidemiol Biomarkers Prev 11: 215-216.
- Ralph DA, Zhao LP, Aston CE, Manjeshwar S, Pugh TW, et al. (2007) Age-specific association of steroid hormone pathway gene polymorphisms with breast cancer risk. Cancer 109: 1940-1948.
- Chen C, Sakoda LC, Doherty JA, Loomis MM, Fish S, et al. (2008) Genetic variation in CYP19A1 and risk of breast cancer and fibrocystic breast conditions among women in Shanghai, China. Cancer Epidemiol Biomarkers Prev 17: 3457-3466.
- Zhang L, Gu L, Qian B, Hao X, Zhang W, et al. (2009) Association of genetic polymorphisms of ER-alpha and the estradiol-synthesizing enzyme genes CYP17 and CYP19 with breast cancer risk in Chinese women. Breast Cancer Res Treat 114: 327-338.
- Iwasaki M, Hamada GS, Nishimoto IN, Netto MM, Motola J, et al. (2010) Dietary isoflavone intake, polymorphisms in the CYP17, CYP19, 17beta-HSD1, and SHBG genes, and risk of breast cancer in case-control studies in Japanese, Japanese Brazilians, and non-Japanese Brazilians. Nutr Cancer 62: 466-475.
- Yoshimoto N, Nishiyama T, Toyama T, Takahashi S, Shiraki N, et al. (2011) Genetic and environmental predictors, endogenous hormones and growth factors, and risk of estrogen receptor-positive breast cancer in Japanese women. Cancer Sci 102: 2065-2072.
- Clendenen T, Zeleniuch-Jacquotte A, Wirgin I, Koenig KL, Afanasyeva Y, et al. (2013) Genetic variants in hormone-related genes and risk of breast cancer. PLoS One 8: e69367.
- Iwasaki M, Mizusawa J, Kasuga Y, Yokoyama S, Onuma H, et al. (2014) Green tea consumption and breast cancer risk in Japanese women: a case-control study. Nutr Cancer 66: 57-67.
- Ghisari M, Eiberg H, Long M, Bonefeld-Jorgensen EC (2014) Polymorphisms in phase I and phase II genes and breast cancer risk and relations to persistent organic pollutant exposure: a case-control study in Inuit women. Environ Health 13: 19.
- Zins K, Mogg M, Schneeberger C, Abraham D, Schreiber M (2014) Analysis of the rs10046 polymorphism of aromatase (CYP19) in premenopausal onset of human breast cancer. Int J Mol Sci 15: 712-724.
- Yang L, Wang XY, Li YT, Wang HI, WU T, et al. (2015) CYP19 gene polymorphisms and the susceptibility to breast cancer in Xinjiang Uigur women. Genet Mol Res 14: 8473-8482.
- Farzaneh F, Noghabaei G, Barouti E, Pouresmaili F, Jamshidi J, et al. (2016) Analysis of CYP17, CYP19 and CYP1A1 Gene Polymorphisms in Iranian Women with Breast Cancer. Asian Pac J Cancer Prev 17: 23-26.
- Kopp TI, Jensen DM, Ravn-Haren G, Cohen A, Sommer HM, et al. (2016) Alcohol-related breast cancer in postmenopausal women - effect of CYP19A1, PPARG and PPARGC1A polymorphisms on female sex-hormone levels and interaction with alcohol consumption and NSAID usage in a nested case-control study and a randomised controlled trial. BMC Cancer 16: 283.
- Ghisari M, Long M, Roge DM, Olsen J, Bonefeld-Jorgensen EC (2017) Polymorphism in xenobiotic and estrogen metabolizing genes, exposure to perfluorinated compounds and subsequent breast cancer risk: A nested case-control study in the Danish National Birth Cohort. Environ Res 154: 325-333.
- Hirose K, Matsuo K, Toyama T, Iwata H, Hamajima N, Tajima K (2004) The CYP19 gene codon 39 Trp/Arg polymorphism increases breast cancer risk in subsets of premenopausal Japanese. Cancer Epidemiol Biomarkers Prev 13: 1407-1411.
- Sobczuk A, Romanowicz H, Fiks T, Polac I, Smolarz B (2009) The CYP17 and CYP19 gene single nucleotide polymorphism in women with sporadic breast cancer. Pol J Pathol 60: 163-167.
- Tuzuner BM, Ozturk T, Kisakesen HI, Ilvan S, Zerrin C, et al ( 2010) CYP17 (T-34C) and CYP19 (Trp39Arg) polymorphisms and their cooperative effects on breast cancer susceptibility. In Vivo 24: 71-74.
- Ramalhinho AC, Fonseca-Moutinho JA, Breitenfeld Granadeiro LA (2012) Positive association of polymorphisms in estrogen biosynthesis gene, CYP19A1, and metabolism, GST, in breast cancer susceptibility. DNA Cell Biol 31: 1100-1106.
- Surekha D, Sailaja K, Rao DN, Padma T, Raghunadharao D, Vishnupriya S (2014) Association of CYP19 polymorphisms with breast cancer risk: A case-control study. J Nat Sci Biol Med 5: 250-254.
- Boone SD, Baumgartner KB, Baumgartner RN, Connor AE, Pinkston CM, et al (2014) Associations between CYP19A1 polymorphisms, Native American ancestry, and breast cancer risk and mortality: the Breast Cancer Health Disparities Study. Cancer Causes Control 25: 1461-1471.
- Alanazi M, Alabdulkarim HA, Shaik JP, Al Naeem A, Elrobh M, et al (2015) No associations between aromatase gene polymorphisms and breast cancer risk in Saudi patients. Onco Targets Ther 8: 2453-2459.
- Baravalle R, Di Nardo G, Bandino A, Barone I, Catalano S, et al. (2017) Impact of R264C and R264H polymorphisms in human aromatase function. J Steroid Biochem Mol Biol 167: 23-32.
- Pan Z, Trikalinos TA, Kavvoura FK, Lau J, Ioannidis JP (2005) Local literature bias in genetic epidemiology: an empirical evaluation of the Chinese literature. PLoS Med 2: e334.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi