Research Article, J Immunol Tech Infect Dis Vol: 12 Issue: 4
In Silico Protein Ligand Docking to Study the Effect of Astaxanthin and Colchicine against Human Papilloma Virus Proteins
Rajarshi Das* and Seneha Santoshi
1Department of Biotechnology, Amity University, Noida, India
*Corresponding Author: Rajarshi Das,
Department of Biotechnology, Amity
University, Noida, India
E-mail: herrrajarshidas@gmail.com
Received date: 01 September, 2023, Manuscript No. JIDIT-23-112002;
Editor assigned date: 04 September, 2023, PreQC No. JIDIT-23-112002 (PQ);
Reviewed date: 18 September, 2023, QC No. JIDIT-23-112002;
Revised date: 25 September, 2023, Manuscript No. JIDIT-23-112002 (R);
Published date: 03 October, 2023, DOI: 10.4172/2329-9541.1000351
Citation: Das R and Santoshi S (2023) Protein Ligand Docking to Study the Effect of Astaxanthin and Colchicine against Human Papilloma Virus Proteins. J Immunol Tech Infect Dis 12:3.
Abstract
Human papilloma virus is a deadly virus that causes cervical, penile and vulval cancer. The current effort focuses on HPV's lesser-known proteins, as well as its virulence and prevalence in men and women. The leitmotif of this study is to create drugs against the proteins of the Human papilloma Virus as a suggestive solution against the diseases caused by this virus. The study includes the extraction of the protein sequences of the virus from NCBI, followed with the secondary structure prediction with the aid of the usage of online available protein prediction tools; GOR4, Chou, Fasman and Phyre2. Homology modelling using SWISSMODEL was done of each protein (E1, E2, L1 and L2) taken from HPV for receiving the appropriate model for the respective proteins. The PDB files for each of the protein were downloaded and used for docking against respective ligands mentioned in this review. The docking results are elaborately discussed in this study.
Keywords
Human Papilloma Virus (HPV); Benign papilloma; E1 protein; E2 protein; L1 protein; L2 protein; Human Papillomaviruses5 Upstream Regulatory Region (HPV5 URR); Q mean; Z score analysis; Astaxanthin; Colchicine; Suberoylanilide Hydroxamic Acid (SAHA)
Introduction
Human Papilloma Viruses (HPV) is non-enveloped tumor viruses with double-stranded DNA of eight kilobases. The viral DNA is encased in a spherical capsid with icosahedral symmetry and a diameter of 1 mm. Around 55 nanometers there are about 100 different varieties of HPV. They infect the people. Benign papillomas are caused by squamous epithelia of the skin and mucosa alternatively, warts. Persistent infection with high-risk oncogenic HPV causes all cervical cancers, cancers of the vulvar, vaginal, penile, and oropharyngeal mucosa, as well as a subset of cancers of the vulvar, vaginal, penile, and oropharyngeal mucosa. Cutaneous beta-HPV variants have been associated to a number of diseases in recent years. Several disorders have been related to cutaneous beta-HPV strains in recent years concerning the pathophysiology of skin tumors that aren't melanoma. For prophylaxis, two HPV vaccines based on Virus-Like Particles (VLP) have been authorized women who have never been infected with HPV 16 or HPV 18; they are up to 100 percent effective at preventing infections and genital lesions. Vaccination prevented HPV 6 and HPV 11 genital warts [1].
HPV is responsible for around 5.2 percent of all cancers worldwide. It’s also linked to a subgroup of anogenital, head, and neck malignancies, as well as cervical cancer. Cervical cancer contributes most to cancer mortality to women worldwide. The variant of HPV highly responsible for cervical cancer are high-risk HPV-16 and HPV-18, which account for over 62.6 percent and 15.7 percent of cervical cancer cases, respectively. Carrageenan prevents the binding of the HPV virions to the cells; it blocks HPV infection through a second, post attachment heparin Sulfate-independent effect. Withaferin a down regulates the expression of HPV E6 oncoproteins and restore the p53 pathway, which results in the apoptosis of cervical cancer cell [1].
Curcumin is cytotoxic and its cytotoxicity is higher in HPV infected cells and it down regulates serine Kinase AKT/nuclear factor κ B pathway via increasing cancer cell sensitivity and regulating HPV oncoproteins [2].
The HPV E7 protein is an oncoprotein that is multi-functional in nature. It induces terminally differentiated cells to enter into the cell cycle. The product of the Retinoblastoma Susceptibility Locus (pRb) and the associated pocket protein family members p107 and p130 interact with this tiny nuclear phosphor protein. In cervical cancer, the interaction of E7 and pRb is critical for transformation and abrogation of anti-proliferative signals [3].
Presence of HPV is insufficient to cause cervical cancer all by itself, as has been proposed by International Agency for Research on Cancer [4].
Papillomaviruses have been formerly grouped with polyomaviruses and simian vacuolating virus within the Papovaviridae own family, however, according to the International Committee on Virus Taxonomy, they are now classified a different family of Papillomaviridae.
In skin, HPV are mainly accountable for benign lesions which broaden on fingers and soles. Various clinical presentations are described, consisting of common warts, filiform warts, flat warts, butchers' warts, mosaic warts and myrmecia. Treatment of warts is frequently hard and includes exclusive unfavorable strategies. Recurrences are frequent because of HPV contamination patience in peri-lesioned skin. Despite the fact that the causal relationship between genital papillomavirus infection and cervical neoplasia has been established, further study is needed to explain and comprehend the implications of HPV in skin cancer [5].
In the United States, the human papillomavirus is the most common sexually transmitted infection, with roughly 80% of women infected by the age of fifty. Despite the fact that most HPV infections are harmless, chronic HPV contamination is linked to the risk of cervical cancer and genital warts. The presently approved quadrivalent HPV vaccination (types 6, 11, 16, and 18) targets the HPV strains responsible for around 70% of cervical malignancies and 90% of genital warts. Cervical Intraepithelial Neoplasia (CIN) grades I, II, and III are all examples of HPV-associated diseases. Cervical Intraepithelial Neoplasia (CIN) grades I, II, and III include Adenocarcinoma In Situ (AIS), vulvar and vaginal neoplasia, and genital warts. The vaccine is normally advised for all girls aged 11 to twelve, with catch-up immunization for females as old as 26, and the vaccine is covered by most insurance policies. A second bivalent HPV vaccination is now awaiting FDA clearance in the United States (FDA). HPV vaccine lowers the prevalence of HPV-related malignancies and precancerous lesions in the United States and overseas, while implementation options are still being considered [6].
High-risk HPV strains, which are now well recognized as the causative agents, cause cervical dysplasia and cancer [7]. According to the American Cancer Society 11,150 new cases of invasive cervical cancer were detected in the United States in 2007, resulting in 3670 fatalities [8]. Despite the fact that, thanks to Papanicolaou testing, cervical cancer has risen to become the second top cause of cancerrelated mortality in women in the United States during the previous 50 years, it remains the world's second leading cause of cancer-related death in women. Invasive cervical cancer affects 510,000 women globally each year, resulting in 288,000 deaths, with around 80% of cases happening in developing nations [9]. In addition to cervical cancer, HPV has been related to other less common genital malignancies such as vulvar, vaginal, and anal carcinomas [10].
Literature Review
In the United States, the most common sexually transmitted infection is the human papillomavirus infection is connected to the development of cervical cancer and genital warts in a major way. The quadrivalent HPV vaccine, which was just approved, targets the HPV strains that cause around 70% of cervical cancers and 90% of genital warts. It can also help to reduce the incidence of HPV-related diseases, especially if administered prior to HPV exposure. The vaccination, which is recommended for all girls aged 11 to 12, is covered by most insurance policies, and with catch-up immunization for women up to the age of 26.A second bivalent HPV vaccine is now being evaluated by the US Food and Drug Administration (FDA). HPV vaccination reduces the prevalence of HPV-related malignancies and precancerous lesions in the United States and elsewhere, however vaccine implementation decisions are still being made [11].
Infection with the human papillomavirus is the primary cause of cancer death in women across the world. HPV's life-threatening infection necessitates the development of anti-cancer medications. Various natural chemicals, such as carrageenan, curcumin, epigallocatechin gallate, indole-3-carbinol, jaceosidin, and withaferin, have been employed as a promising source of anticancer treatment in recent years. Several studies have illustrated that these chemicals can prevent HPV infection. We investigated these natural inhibitors against the E6 oncoprotein of high-risk HPV-16, which is known to inactivate the p53 tumor suppressor protein, in the current work. Using a structure-based drug design technique, a robust homology model of HPV-16 E6 was constructed to predict the interaction mechanism of E6 oncoprotein with natural inhibitory compounds. The interaction of these natural chemicals with the p53-binding site of E6 protein residues 113-122 (CQKPLCPEEK) was discovered using docking studies, which aided in the restoration of p53 function. In addition to assisting the in silico validation of natural chemicals, docking analysis aids in the understanding of molecular processes of protein-ligand interactions [12].
The E1 and E2 proteins of the human papillomavirus bind cooperatively to the viral Origin of Replication (ori), generating an E1-E2-ori complex that is required for DNA replication to begin. The host cell provides all other replication proteins, including DNA Polymerase Primase (pol-primase). We investigated the interactions of HPV type 16 (HPV-16) E1 with E2, ori, and the four pol-primase subunits in great detail. E2 binding requires a C-terminal portion of E1 (Amino Acids (aa) 432 to 583 or 617) according to deletion study.HPV-16 in the absence of E2, E1 was unable to bind the ori, but E1's C-terminal domain was sufficient to tether E1 to the ori via E2. Only p68 bound E1 among the pol-primase subunits, and binding was competitive with E2. The E1 area needed (aa 397 to 583) was the same as for E2 binding, but with the addition of 34 N-terminal residues. We discovered that a monoclonal antibody mapped close to the N-terminal junction of the p68-binding domain prevented E1-p68 but not E1-E2 binding, confirming these distinctions. The mapping data and sequence alignments for HPV-16 E1 and other Superfamily 3 (SF3) viral helicases show that aa 439 to 623 form a distinct helicase domain. We created a structural model of this domain based on the Xray structures of the hepatitis C virus and Bacillus stearothermophilus (SF2) helicases, assuming a similar nucleoside triphosphate-binding fold. Our findings imply that this area of E1 is multifunctional and crucial to various phases of HPV DNA replication, since the modelling closely fits the deletion study [13].
Human papillomavirus infection causes cervix cancer, which is the second leading cause of mortality in women globally. The binding site of HPV 16 E1-E2 protein has yet to be determined from a 3-D structure, thus we used biphenylsulfonacetic acid as an inhibitor to investigate the structure of E1-E2 protein from Human papillomavirus type 16 and uncover possible binding sites. For 3D structure prediction, the Swiss model was employed, and PDB: 2V9P (E1 protein) and 2NNU (E2 protein) with 52.32 percent and 100 percent similarity, respectively, were chosen as templates. E1 and E2 created a 3D model structure that was 99.2 percent and 99.5 percent in the core and permitted regions, respectively. The ligand binding sites were predicted with the internet server Meta pocket 2.0, and docking was done with MOE 2009.10. HPV-16's E1 and E2 proteins have three putative binding sites that might interact with inhibitors. The ligand interacts with the protein by hydrogen bonds on Lys 403, Arg 410, His 551 in the first pocket, Tyr 32, Leu 99 in the second pocket, and Lys 558 Lys 517 in the third pocket, according to the docking biphenylsulfonacetic acid utilising these binding sites [14].
Human papillomavirus genome replication necessitates the use of E1 and E2 proteins as viral trans-factors and the replication origin, which is situated in the URR, as a cis-element. The minimum requirements for an HPV replication origin vary depending on the virus type, but they always involve one or more E2 protein binding sites. In contrast to beta-HPV8, the requirements for an E1 binding site appear to vary among HPV genera, with alpha-HPV11 and -18 minimum origins able to replicate without an E1 binding site. The sequencing requirements for the beta-HPV5 minimum origin of replication were investigated in this paper. Given that three E2 binding sites are intact and both viral replication proteins are present, we show that the HPV5 URR may replicate in U2OS cells without the sequence suggested as an E1 binding site, albeit at lower levels than the URR. The lack of an absolute necessity for the E1 binding site for HPV5 replication origin led us to investigate if additional HPV E1 and E2 proteins are capable of supporting replication from this origin. In contrast to proteins from alpha-HPV types 11, -16, or -18, the E1 and E2 proteins from beta-HPV types enable replication from the origin. Furthermore, even if the E1 binding site is intact, the replication proteins E1 and E2 of these alpha-HPV types are unable to sustain the replication of HPV5 URR. Following these findings, we investigated the potential of various combinations of E1 and E2 proteins from various alpha and beta-HPV types to enable the replication of URR sequences from the relevant HPV types in the U2OS cell line [15].
The E2 DNA-binding domain of the high-risk cervical cancerassociated strain Human Papillomavirus Type 16 (HPV-16) has been crystallised and characterised. The E2 proteins of the papillomavirus control transcription from all viral promoters and are needed for replication to begin in vivo. They are members of a family of viral proteins that form dimeric beta-barrels and interact with DNA via surface alpha-helices. Although all E2 proteins identify the same palindromic DNA sequence, the ability of proteins from various viral strains to differentiate between their unique DNA-binding sites varies. While the overall fold of the HPV-16 E2 DNA-binding domain matches that of the related viral strain bovine papillomavirus type 1, the specific positioning of the recognition helices is notably different, as revealed by the structure. Furthermore, the charge distribution on the two proteins' DNA-binding surfaces differs; HPV-16 E2 has a substantially less electropositive surface. As a result, HPV-16 E2 is less able to induce bending by using charge neutralisation of phosphate groups on DNA. These findings are consistent with earlier solution investigations, which found that HPV-16 E2 had a lower affinity for flexible DNA target sequences and a higher affinity for Atract- containing, pre-bent sequences. In conclusion, the crystal structure of the HPV-16 E2 DNA-binding domain reveals that the protein displays a stereochemically and electrostatically distinct interface to DNA, which may play a role in its DNA target discriminating mechanism [16].
Materials and Methods
Methodology of this research involves several steps; most of them have been narrated below.
Retrieving information from databases
National Center for Biotechnology Information (NCBI): From NCBI we have collected the accession number, protein sequence and nucleotide sequence of the four proteins of HPV.
Protein Data Bank (PDB): From PDB we have collected the accession number of proteins along with their sequences.
Swissprot (UniProt): From Swissprot we have collected the accession number of the proteins that is required in our docking research.
Structure prediction and modelling tools
Garnier Osguthorpe Robson (GOR): The GOR technique (short for Garnier-Osguthorpe-Robson) is a prediction approach for secondary structures in proteins based on information theory. We have used GOR software online for secondary structure prediction for the purpose of analysis of the percentage composition of the secondary structures present in each chosen protein of HPV.
Phyre2: Protein Homology/Analogy Recognition Engine; pronounced "fire" (Phyre and Phyre2) are two free web-based protein structure prediction systems. Phyre is one of the most widely used algorithms for predicting protein structure; we have used Phyre2 for protein secondary structure prediction.
Chou-fasman: The Chou-Fasman approach is an empirical method for predicting tertiary structures in proteins. Based on known protein structures determined with X-ray crystallography, the approach analyses the relative frequencies of each amino acid in alpha helices, beta sheets, and twists. Chou-fasman gives us detail about the structure components of the protein model, like the number of turns, coils present etc. We have used Chow-fasman for protein tertiary structure prediction.
Swiss model: SWISS-MODEL is a fully automated protein structure homology-modeling system that may be accessed via the Expasy web server or the Deep View software (Swiss Pdb-Viewer). We have used SWISS-MODEL to find out the model of the structure of each chosen protein of the HPV Virus.
Drug docking
Swiss dock: Swiss Dock is an online tool that forecasts potential molecular interactions between a target protein and a small molecule. We have utilized Swiss dock for performing molecular docking with the protein of interest and a ligand and the results have been recorded.
Results
The present study involves the in-silico analysis of the lesserknown proteins of Human Papillomavirus namely E1, E2, L1, L2. These proteins were identified and their protein sequences were extracted. The study then utilized different Protein structure prediction tools to help analyse the structure. The details obtained through the different tools was compared and analysed.
From literature various different drugs and inhibitors were extracted and docking studies were done using the above four proteins as target proteins (Figure 1).
Protein structure prediction by using structure prediction tools
Different Protein structure prediction tools were used for prediction of the HPV proteins. The use of different tools helped in complete analysis of the structure and a better understanding of the tool’s efficiency.
By using Phyre2: Structure prediction with phyre2 is done to detect the percentage of Alpha helices and Beta sheets.
By using GOR4: GOR4 has been used to predict secondary structure of the protein molecule.
By using Chou-Fasman: Chou and Fasman tool has been used to predict the secondary structure of the protein molecules (Table 1).
| E1 | E2 | L1 | L2 | |
|---|---|---|---|---|
| Phyre | Alpha helix: 39% | Alpha helix: 26% | Alpha helix: 12% | Alpha helix: 6% |
| Beta sheet: 12% | Beta sheet: 22% | Beta sheet: 29% | Beta sheet: 18% | |
| Disordered: 21% | Disordered: 30% | Disordered: 28% | Disordered: 55% | |
| Chou-fasman | Alpha helix: 81.2% | Alpha helix: 60.5% | Alpha helix: 59.8% | Alpha helix: 53.70% |
| Beta sheet: 0.00% | Beta sheet: 17.2% | Beta sheet: 15% | Beta sheet: 16.10% | |
| GOR4 | Alpha helix: 31.67% | Alpha helix: 25.00% | Alpha helix: 21.98% | Alpha helix: 21.63% |
| Beta sheet: 0.00% | Beta sheet: 0.00% | Beta sheet: 0.00% | Beta sheet: 0.00% | |
| Random coiled: 47.33% | Random coiled: 55.75% | Random coiled: 57.30% | Random coiled: 59.20% |
Table 1: Structural differences of the different proteins of HPV as shown in different structure predicting tools.
Homology model making by use of Swiss Modeller
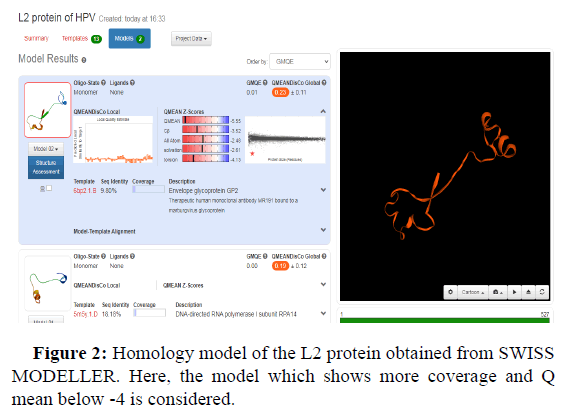
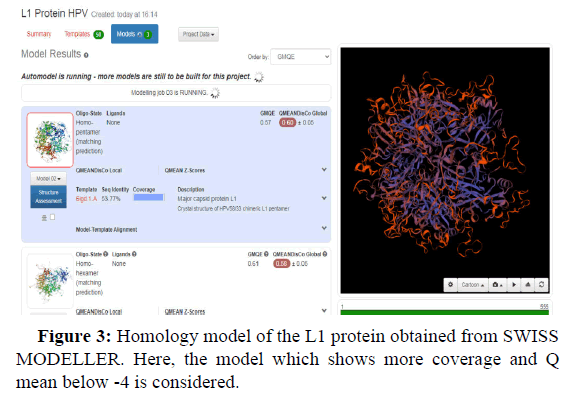
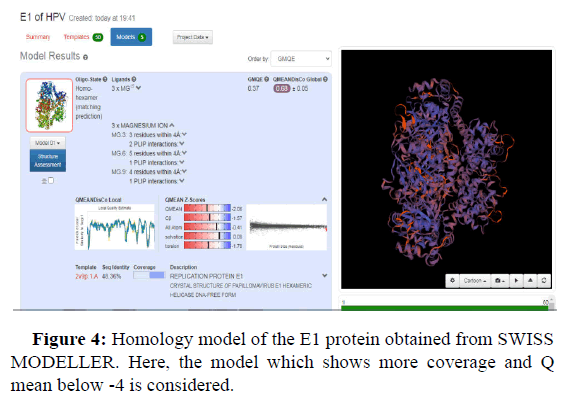
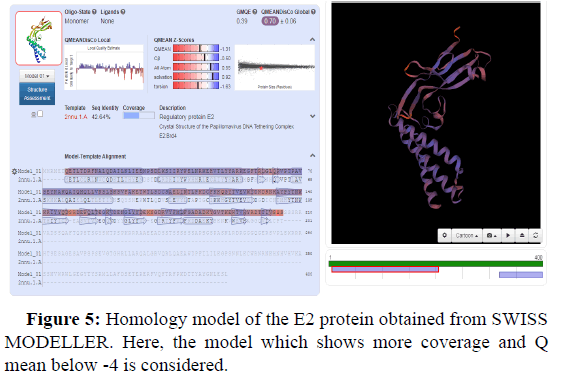
Swiss model is an in silico tool which gives the best threedimensional structure of proteins. Its prediction is based on homology modelling (Figures 2-5).
GMQE and QMEANDisCo global values in the Swiss model provide a quality measurement data between 0 and 1. This means that values nearer 1 will have a better structure prediction. GMQE value is coverage dependent that means that if the structure predicted is for total protein sequence, then values will be higher than 0.5-0.6. But QMEANDisCo values are not dependent on the sequence coverage.
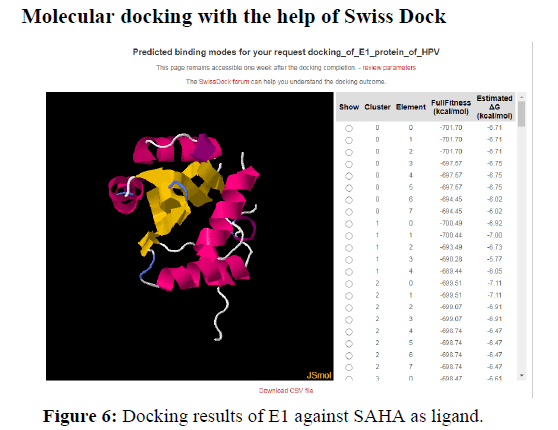
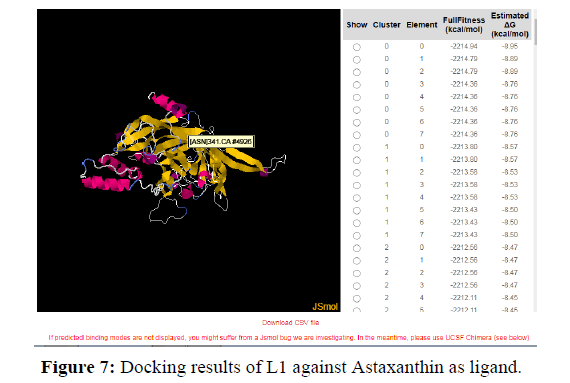
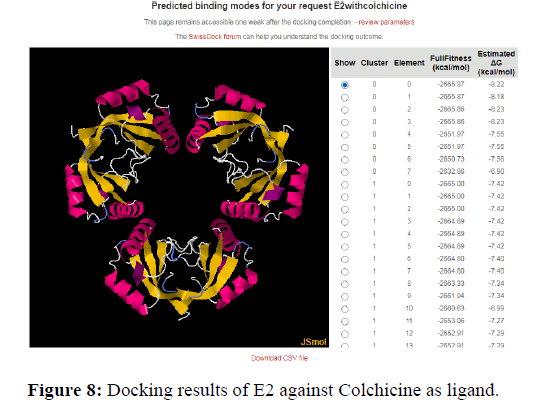
Another value known as QMEAN Z-score analysis is also used for global model quality estimates. It is based on 4 statistical potentials of mean force and their linear combination: the "QMEAN" score. In other words: Z-scores around 0.0 would provide a “native-like" structure and, but a "QMEAN" Z-score below -4.0 indicates a model with low quality (Figures 6-8).
Molecular docking with the help of Swiss Dock
The docking analysis shows that Colchicine and Astaxanthin give a good binding or docking results to the HPV proteins. These drugs or inhibitors can thus be used for stopping the progression of the virus in human cells. Higher the negative values for delta G higher is the binding capacity and the fitness of docking.
Discussion
Protein structure Prediction provides an understanding of how a protein works, which allows the researchers to create hypotheses about how to affect it, control it, or modify it. Thus, analyzing a protein's structure allows us to design site-directed mutations with the intent of changing the function [17].
The Protein structure depends on its amino acid sequence and local, low-energy chemical bonds between atoms in both the polypeptide backbone and in amino acid side chains. As it is very precisely known that the protein structure plays a key role in its function and if a protein loses its shape at any structural level, it may no longer be functional or active. Modelling means constructing an atomicresolution model of the "target" protein from its amino acid sequence after comparing it with the structures available in the data.
Swiss Model is a fully automated protein structure homologymodelling server, accessible via the Expasy web server. The purpose of this server is to make protein modelling accessible to all life science researchers worldwide. Swiss model calculates the QMEAN, which stands for Qualitative Model Energy Analysis. It’s a scoring function that describes the key geometrical properties of protein structures as a whole. A torsion angle potential is used to examine the local geometry over three successive amino acids. In SWISS-MODEL the QMEAN zscore represents an estimate of how comparable the model is to experimentally derived structures of similar size. QMEAN z-scores around zero indicate good agreement between the model structure and experimental structures of similar size. In most of the Swiss models predicted the QMEAN is between 0-1. This shows that a stable and functional model has been predicted.
PHYRE 2 structure prediction tool has been used to analyses and understand the Secondary structure and disorder prediction. The prediction is 3-state: α-helix, β-strand or coil. α-helices are shown by green helices, strands are represented by blue arrows, and coil is represented by faint lines. The 'SS confidence' line shows how confident the forecast is, with red indicating high confidence and blue indicating low confidence.
The 'Disorder' line comprises the protein's predicted disordered sections, which are denoted by question marks (?). The weakest region of helix prediction coincides with a relatively strong prediction of disorder. Disordered regions can often by functionally very important and because they are disordered, their structure is not required to be predicted and it can keep changing.
Docking analysis
The molecular docking studies were done using an online tool SWISS DOCK. Docking was done to get Target protein Drug Interaction between HPV proteins and four drugs. The antiviral drugs taken for docking include:
• SAHA
• Astaxanthin
• Colchicine
A binding between a protein and Ligand is better when there is less spontaneity and more stability. So, the docking results show that drugs gave a docking score of -7.1 and -8.4 with HPV proteins. Thus, higher the negative score better is the binding and stability of docking. This negative energy results are nothing but the Gibb’s free energy. According to the principles and fundamentals of thermodynamics, we know that Gibb’s free energy is inversely proportional to the spontaneity of a particular reaction. Therefore, more negative the Gibb’s free energy is the more stable the binding is.
Conclusion
By computationally generating and analyzing protein structures from amino acid sequences, protein structure prediction and modelling creates tools for researching the foundation of protein activities. Users can access the information they need using protein prediction tools and web servers. Models of three-dimensional protein structures and learn about the activities of various proteins. Protein structure prediction and modelling will be important in the age of precision medicine. Increasingly significant roles in the study of illness biological mechanisms and drug development design. Drugs that can bind to the target ligand can be found once the target protein has been identified and its structure predicted using bioinformatics methods. This may be accomplished via Molecular Docking, which can be done using a variety of servers or programs, such as SWISS DOCK, 1-Click docking, and so on are available on the internet. This is the current study's goal, thus it's critical to find out more about it. Many dangerous illnesses have been cured. The purpose of this study is to locate and download the Gene and its variants. Proteins that play a role in human disorders such as cervical cancer SWISS PROT/PDB may also be used to predict the structure of proteins, receptors, or membrane proteins, as well as find ligands and inhibitors and their structures. For drug discovery, docking investigations utilizing the Swiss Dock tool are used. In drug development and prediction, which can bind to target ligand has been found to be highly useful.
References
- Handisurya A, Schellenbacher C, Kirnbauer R (2009) Diseases caused by human papillomaviruses (HPV). J Dtsch Dermatol Ges 7(5):453-466.
- Kumar S, Jena L, Sahoo M, Kakde M, Daf S, et al. (2015) In silico docking to explicate interface between plant-originated inhibitors and E6 oncogenic protein of highly threatening human papillomavirus 18. Genomics Inform 13(2):60.
- Yim EK, Park JS (2005) The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat 37(6):319-324.
- Meites E, Gee J, Unger E, Markowitz L (2020) Human Papilloma Virus. Centers for disease control and prevention (CDC).
- Aubin F, Laurent R (2006) Human papillomavirus-associated cutaneous lesions. Rev Prat 56(17):1905-1913.
- Braaten KP, Laufer MR (2008) Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev Obstet Gynecol 1(1):2.
- Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV (2002) The causal relation between human papillomavirus and cervical cancer. J Clin Pathol 55(4):244-265.
- Atlanta GA (2007) Cancer Facts and Figures 2007. American Cancer Society
- World Health Organization (WHO) (2008) “Human Papilloma Infection and Cervical Cancer”.
- Hildesheim A, Han CL, Brinton LA, Kurman RJ, Schiller JT (1997) Human papillomavirus type 16 and risk of preinvasive and invasive vulvar cancer: Results from a seroepidemiological case-control study. Obstet Gynecol 90(5):748-754.
- Braaten KP, Laufer MR (2008) Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev Obstet Gynecol 1: 2
- Kumar S, Jena L, Galande S, Daf S, Mohod K, et al. (2014) Elucidating molecular interactions of natural inhibitors with HPV-16 E6 oncoprotein through docking analysis. Genomics inform 12: 64
- Masterson PJ, Stanley MA, Lewis AP, Romanos MA (1998) A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase α-primase p68 subunit. J Virol 72: 7407-19
- Iryani I, Amelia F, Iswendi I (2018) Active sites prediction and binding analysis E1-E2 protein human papillomavirus with biphenylsulfonacetic acid. IOP Conf Ser Mater Sci Eng 335(1):012031
- Laaneväli A, Ustav M, Ustav E, Piirsoo M (2019) E2 protein is the major determinant of specificity at the human papillomavirus origin of replication. PloS one 14: e0224334
- Hegde RS, Androphy EJ (1998) Crystal structure of the E2 DNA-binding domain from human papillomavirus type 16: Implications for its DNA binding-site selection mechanism. J Mol Biol 284:1479-89
- Altschul SF (2007) Sense from Sequences on Bettering BLAST. J Mol Biol 215: 403-10
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi