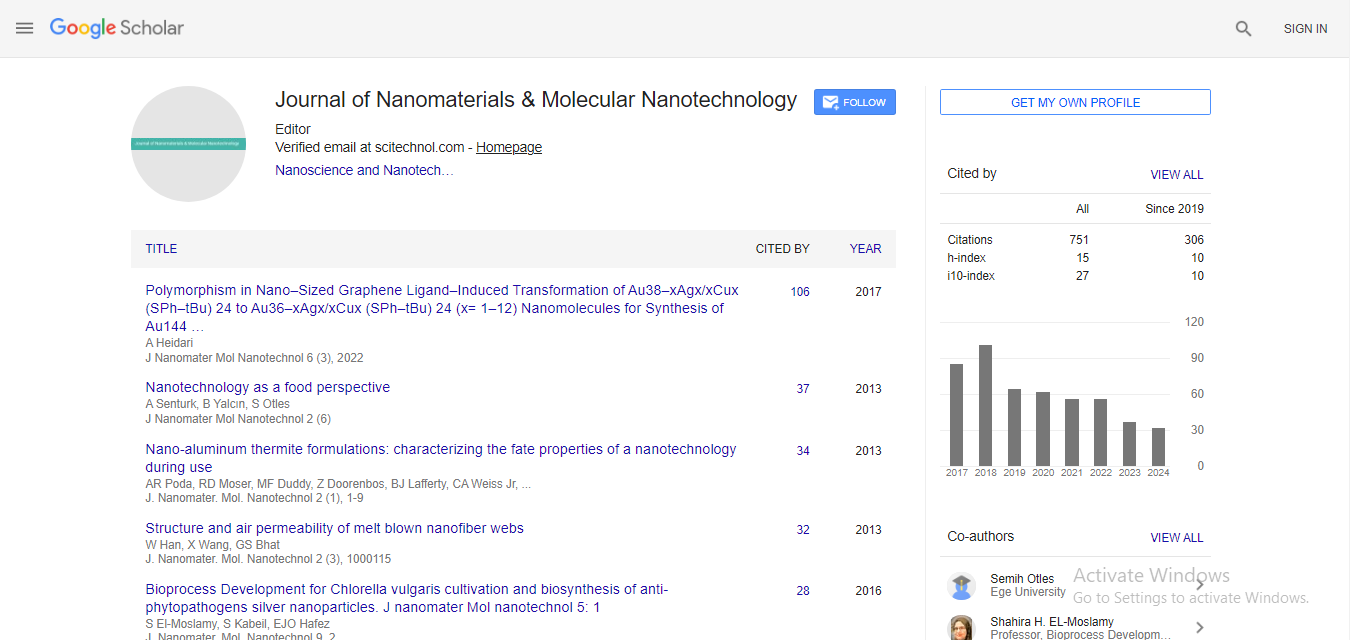
Solute binding proteins and their cognate ligands: structure, function and their role in functional annotation
Umesh Yadava
Deen Dayal Upadhyaya Gorakhpur University, India
: J Nanomater Mol Nanotechnol
Abstract
The uptake of exogenous solutes is mediated by transport systems embedded in the plasma membrane and drive active transport even at μM to nM solute concentrations. In many of these systems, a periplasmic Solute-Binding Protein (SBP) is utilized to bind their cognate ligands with high affinity and deliver them to the membrane-bound translocator subunits. Knowledge of the cognate ligand for the SBP component of the transporter can provide crucial data for functional assignment of co-located or co-regulated genes. In the present study, the structural and functional characterizations of several solute binding proteins have been carried out. Proteins were cloned from genomic DNA, expressed by autoinduction, and purified by a combination of Ni-NTA and size exclusion chromatography. The purified SBPs were screened using differential scanning fluorimetry (DSF) and about 400 compounds ligand library. Two of the SBPs exhibited DSF hits that were novel for their respective transport family. Crystallization trials of proteins have been conducted with their respective DSF ligand hits. Those SBPs that have structures determined and their respective interactions with co-crystallized ligands will be presented. Co-crystallization with DSF determined ligands resulted in structures of Avi_5305 in complex with D-glucosamine and D-galactosamine, the first structure of an ABC SBP with an amino sugar. Further, for the study of detailed interactions at the binding site and their timedependent behavior, the docking of ligands within the respective binding sites and molecular dynamics simulation of the best-docked poses have also been performed. Energy values and RMS deviation plots during the course of dynamics and other thermodynamic parameters indicate the better binding capabilities and stabilities of the ligands within active sites.
Biography
Prof. Umesh Yadava has completed his PhD from DDU Gorakhpur University, Gorakhpur, India and postdoctoral studies from Albert Einstein College of Medicine, New York, USA. Currently, he is working as Professor in the Department of Physics, DDU Gorakhpur University, Gorakhpur. He is the recipient of Raman Fellowship and Young Scientist awarded by University Grants Commission, New Delhi and Department of Science and Technology, New Delhi, India respectively. He has published more than 45 papers in reputed journals and has been serving as an editorial board member of repute. (https://orcid.org/0000- 0002-9127-532X)
E-mail: u_yadava@yahoo.com
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi