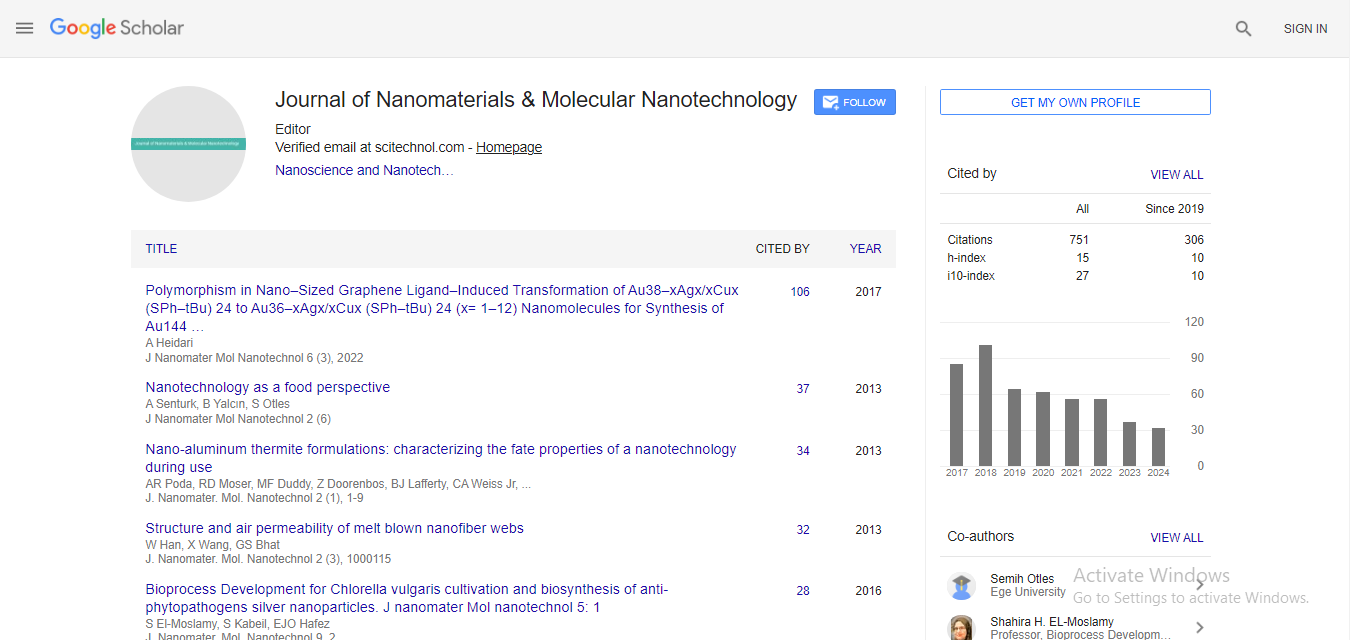
Commentary, J Nanomater Mol Nanotechnol Vol: 12 Issue: 6
Applications and Advancements in Molecular Dynamics Simulation
Jaursup Youngme*
1Department of Chemistry, Khon Kaen University, Khon Kaen, Thailand
*Corresponding Author: Jaursup Youngme,
Department of Chemistry, Khon
Kaen University, Khon Kaen, Thailand
E-mail: youngmejaursup@gmail.com
Received date: 20 November, 2023, Manuscript No. JNMN-23-123833;
Editor assigned date: 22 November, 2023, Pre QC No. JNMN-23-123833 (PQ);
Reviewed date: 06 December, 2023, QC No. JNMN-23-123833;
Revised date: 13 December, 2023, Manuscript No. JNMN-23-123833 (R);
Published date: 20 December, 2023, DOI: 10.4172/2324-8777.1000386
Citation: Youngme J (2023) Applications and Advancements in Molecular Dynamics Simulation. J Nanomater Mol Nanotechnol 12:6.
Description
Molecular Dynamics Simulation (MDS) stands at the forefront of modern computational chemistry, offering a powerful tool for researchers to unravel the dynamic behavior of molecules at the atomic and molecular levels. This computational technique provides valuable insights into the complex and intricate world of biomolecules, materials, and chemical reactions. By numerically solving Newton's equations of motion, MDS allows scientists to simulate the temporal evolution of a system, capturing details that are challenging or impossible to observe experimentally.
Principles of molecular dynamics simulation
At its core, Molecular Dynamics Simulation is based on classical mechanics, utilizing Newton's laws of motion to describe the movement of atoms and molecules. The system under investigation is represented as a set of interacting particles, each possessing specific properties such as mass, charge, and van der Waals forces. The simulation proceeds by integrating the equations of motion over tiny time steps, calculating the positions and velocities of particles at each step. The result is a trajectory that provides a detailed account of the molecular motions over time.
The force field, a mathematical model that quantifies the interactions between particles, plays a pivotal role in MDS. Force fields include parameters that dictate the strength and nature of various forces, such as bond stretching, angle bending, and non-bonded interactions. The accuracy of a simulation heavily relies on the appropriateness of the chosen force field, and researchers often finetune parameters based on experimental data.
Applications of molecular dynamics simulation
MDS finds applications across diverse scientific domains, ranging from biology and chemistry to materials science and nanotechnology.
In the area of biomolecular simulations, researchers employ MDS to study protein folding, investigate drug-protein interactions, and explore the dynamics of nucleic acids. By simulating the behavior of these macromolecules, scientists gain insights into their function, stability, and response to external stimuli.
In materials science, Molecular Dynamics Simulation is a valuable tool for exploring the mechanical, thermal, and electronic properties of materials. Researchers can investigate the behavior of materials under different conditions, such as extreme temperatures or pressures, providing critical information for designing new materials with specific functionalities.
Chemical reactions are another area where MDS excels. By simulating the trajectory of reacting molecules, researchers can elucidate reaction mechanisms, identify intermediate states, and understand the factors influencing reaction rates. This information is vital for designing catalysts and optimizing reaction conditions in chemical processes.
Advancements in molecular dynamics simulation
Over the years, advancements in computational power and algorithmic techniques have propelled Molecular Dynamics Simulation to new heights. High-performance computing clusters and specialized hardware, such as Graphics Processing Units (GPUs), enable researchers to simulate larger and more complex systems with unprecedented accuracy and efficiency.
Enhancements in sampling methods, such as replica exchange and metadynamics, address the challenge of exploring the vast conformational space of biomolecules. These techniques facilitate the simulation of rare events, allowing researchers to capture transitions between different states and uncover dynamic phenomena that were once elusive.
Machine learning techniques are increasingly integrated into MDS to enhance the accuracy of force fields and improve the efficiency of simulations. By training models on extensive datasets, machine learning algorithms can learn intricate patterns in molecular interactions, leading to more realistic simulations and faster convergence.
Molecular Dynamics Simulation has evolved into a cornerstone of modern computational chemistry, empowering researchers to explore the dynamic behavior of matter at the atomic and molecular scales. From unraveling the secrets of biomolecular interactions to designing advanced materials with tailored properties, MDS has become an indispensable tool in the scientist's toolkit. As computational capabilities continue to advance, the future holds the promise of even more realistic and insightful simulations, further expanding our understanding of the intricate dance of atoms and molecules.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi