Review Article, La Prensa Medica Vol: 111 Issue: 1
Dysregulated lncRNAs in Mesothelioma and their Potential Use in Diagnosis: A Systematic Review
Dina M Elkahwagy1, Caroline Joseph Kiriacos1 and Manar Mansour2*
1Department of Pharmaceutical Biology, German University in Cairo, Cairo, Egypt
2Department of Pharmaceutical Biology and Microbiology, German University in
Cairo, Cairo, Egypt
*Corresponding Author:Manar Mansour
Department of Pharmaceutical Biology and Microbiology, German University in Cairo, Cairo, Egypt
E-mail: manar.soliman@guc.edu.eg
Received date: 04 December, 2023, Manuscript No. IPMA-23-122010;
Editor assigned date: 07 December, 2023, PreQC No. IPMA-23-122010 (PQ);
Reviewed date: 21 December, 2023, QC No. IPMA-23-122010;
Revised date: 06 February, 2025, Manuscript No. IPMA-23-122010 (R);
Published date: 13 February, 2025, DOI: 10.4172/0032-745X.1000685
Citation: Elkahwagy DM, Kiriacos CJ, Mansour M (2025) Dysregulated lncRNAs in Mesothelioma and their Potential Use in Diagnosis: A Systematic Review. La Prensa Medica 111:1.
Abstract
Background: Malignant mesothelioma is a rare and aggressive form of cancer caused by uncontrolled growth and proliferation of pleural mesothelial cells. Mesothelioma is often diagnosed at an advanced stage, leading to significant challenges in treatment and prognosis. The absence of a reliable and routine screening test for mesothelioma further contributes to late diagnosis. The quest for new diagnostic tools imposes a need to improve early detection and enhance patient outcomes. Biomarkers are biological indicators of a biological state or disease. Long noncoding RNAs (lncRNAs) have emerged as potential diagnostic markers for mesothelioma. Researchers have identified specific lncRNAs that show differential expression patterns in mesothelioma tissues compared to healthy tissues, making them attractive candidates for diagnostic purposes.
Aim: Our aim is to assess the potential of using lncRNAs as diagnostic biomarkers in mesothelioma. Data extraction will be performed to collect information on the different lncRNAs studied, their expression patterns, and diagnostic accuracy measures. We will also analyze the methodologies employed for lncRNA detection and quantification. This review will follow a rigorous and transparent methodology, adhering to established guidelines for systematic reviews.
Methods: In this review, we systematically searched multiple databases to identify relevant studies that investigated lncRNAs as diagnostic markers in mesothelioma. Four databases were used in our search, namely, PubMed, JStor, Mdpi and ScienceDirect, for articles published between 2010 and 2022. The search criteria were based on the keywords “mesothelioma”, “lncRNA” or “long noncoding RNA”. The study was independently evaluated by 2 researchers. After screening 1141 articles, only 3 articles were included for fulfilling the criteria. Quality assessment was carried out using Diagnostic Accuracy Studies (QUADAS)-2
Results: Three studies analyzed and confirmed the expression and diagnostic value of four lncRNAs (GAS5, lncRNA?RP1 86D1.3, SNHG8, POT1-AS1) by RT?qPCR in 96 patients of various ethnic backgrounds and gender groups.
Conclusion: The findings of this systematic review will contribute valuable insights to the field, inform future research directions, and potentially guide the development of lncRNA based diagnostic approaches for mesothelioma.
Keywords: Mesothelioma, Malignant pleural mesothelioma, Long noncoding RNA, LncRNA, Diagnostic biomarker, Systematic review
Keywords
Mesothelioma; Malignant pleural mesothelioma; Long noncoding RNA; LncRNA; Diagnostic biomarker; Systematic review
Introduction
Sequencing of the human genome has recently revealed that only approximately 1-2% of the genes present code for proteins, and new tools such as tiling resolution genomic microarrays and whole genome and transcriptome sequencing technologies have enabled detailed characterization of these sequences. The rest were later determined to code for noncoding RNAs (ncRNAs); those sequences were initially believed to be only transcriptional ‘debris’; byproducts with no functional use. Recent accumulating evidence has shown that a growing number of lncRNAs exert cellular regulatory functions [1-3].
Current known classes of noncoding RNAs can be divided according to their length (small or short, 18-200 nucleotides, and long, more than 200 nucleotides) or functionality (housekeeping or regulatory) (Table 1). Crossover of properties exists, so truly discrete categories are difficult to distinguish [4].
| Housekeeping ncRNAs | Regulatory ncRNAs | |
| Long noncoding RNA | Short noncoding RNA | |
| Ribosomal RNAs (rRNAs) Transfer RNAs (tRNAs) Small nuclear RNAs (snRNAs) Small nucleolar RNAs (snoRNAs) |
Long intergenic noncoding RNAs (lincRNAs) Natural antisense transcripts (NATS) Circular RNAs (circRNAs) Pseudogenic transcripts Long enhancer noncoding RNAs (eRNAs) Transcribed ultraconserved regions (T-UCRs) Intronic lncRNA |
microRNAs (miRNAs) PIWI-interacting RNAs (piRNAs) Endogenous small interfering RNAs (siRNAs) Promotor associated RNAs (pRNA) |
Table 1: A summary of one way of classifying noncoding RNAs adapted from. NcRNAs may be classified into 2 major categories according to their functions: Housekeeping and regulatory. The regulatory ncRNAs are further sub-classified into short (less than 200 nucleotides) or long (more than 200 nucleotides), depending on the length of the fragment. Housekeeping RNAs are usually important for cell functions and hence are ubiquitously expressed. Regulatory ncRNAs function as mediators of cellular activities.
LncRNAs
Although no definition exists that is agreed upon by the scientific community, lncRNAs are generally defined as transcripts with a length of more than 200 bp and a lack of translated Open Reading Frames (ORFs) and thus do not code for proteins [5,6]. Exceptionally, recent transcriptomic and proteomic analyses have shown that some lncRNAs do contain short, cryptic ORFs. Moreover, some lncRNAs have been found to be potentially translated into peptides or micro proteins themselves.
As of January 2022, the LNCipedia database website has compiled 56,946 genes and a list of ~127,802 lncRNA transcripts across various species and tissues [7].
The current annotated genes produced by the GENCODE consortium indicate 14,880 transcripts and an estimation of more than 16,000 lncRNA genes within the human genome, with estimates exceeding 100,000 human lncRNAs [8].
The amount of lncRNA present increases with the genome size and the proportion of noncoding DNA, which implies that lncRNA may have developed at a later stage in evolution [9].
Most lncRNAs are structurally similar to mRNAs, as they are transcribed by RNA polymerase II and ENA polymerase III in eukaryotes. The resulting transcribed lncRNAs are further capped by 7-methyl guanosine at the 5’ end and a poly A tail at the 3’ end and finally undergo splicing together [6,10]. Some lncRNAs lack such structures and must be stabilized in other ways, especially at their 3’ ends [9].
A growing body of research has shown that lncRNAs are more than junk RNAs and are instead common in many cellular regulatory functions. The exact nature of the versatile functions still needs to be further elucidated [1]. LncRNAs can have different classifications based on their structural sequence, localization, function, metabolism, or interactions with DNA elements, such as protein-coding genes. There has never been a generalized classification for lncRNAs thus far [11,12].
LncRNAs have been found to regulate gene expression at the epigenetic, transcriptional and posttranscriptional levels through distinct and highly diverse functions [13-15]. LncRNAs may act through four mechanisms: signal, decoy, guide, and scaffold. LncRNAs regulate gene expression through different mechanisms of action and at various levels. While other mechanisms have been proposed, they have not been fully explored. One such example is by acting as enhancer RNA (eRNA) to cause an effect such as enzymatic activity modulation or chromatin looping.
CONCR is one such lncRNA reported to interact with an enzyme called DNA helicase (DDX11) that is involved in DNA replication, organization of heterochromatin and facilitation of sister chromatid cohesion during cell division [16-18].
Some lncRNAs, such as SWINGN, induce chromatin looping to allow Protein-Coding Genes (PCGs) to come into contact with their enhancers if they are close enough or recruit looping factors in distant regions to initiate regions [19].
Some lncRNAs exert their function by acting as molecular scaffolds, creating a center that assembles different proteins to form RNA-protein or multi-protein complexes that then activate or suppress the transcription of genes [20].
LncRNAs may also act as decoys. Regulatory factors such as transcription factors, miRNA or chromatin are falsely sequestered by the decoy, interacting with the decoy lncRNA instead of binding to their intended target sites and thereby regulating gene expression. Linc-MD1 competes as a decoy with the mRNA targets MAML1 and MEF2C for the binding of the regulators, miR-133 and miR-135 to repress the expression of specific genes involved in muscle differentiation.
Moreover, they may act as guides, binding and influencing the movement of factors to precise locations whether close or distant in the genome. The destination of the guide depends on the nature of the interaction itself, whether RNA-RNA, RNA-DNA, or RNA-proteins. For example, the famed HOTAIR may act as a guide by binding and directing Polycomb Repressive Complex 2 (PRC2), a chromatin modifier, to the distant HOXD gene locus and causing an inhibition of its gene expression. PRC2 may be guided by other lncRNAs, such as MEG3 and KCNQ1OT1.
They may also be expressed as signal molecules in response to environmental stimuli at a specific space and time. Thus, their presence may serve as a phenotypical indicator of the transcriptional activity of specific stimuli triggering their production.
Consequently, their involvement in cellular physiology means that any alteration in their expression patterns may actively take part in the pathogenesis of many diseases. In fact, abnormalities in lncRNAs and their expression levels have been found to be directly linked with many human illnesses.
The altered levels of expression may serve as biomarkers for diagnosis, while continuous monitoring of these levels can help with prognosis and can be used in novel therapeutic strategies in the treatment of diseases through targeting pathways they affect.
LncRNAs pioneering as biomarkers
Aberrant lncRNA expression has been observed and validated in many cancers/tumors, which points to potential roles that have not yet been fully understood in cancer biology.
The presence of lncRNAs in clinical samples can serve as molecular biomarkers with diagnostic and prognostic value, and their potential in this avenue has been extensively evaluated and proven in other cancers. LncRNAs exhibit various properties that may enable them to be highly successful as biomarkers. Their wide-ranging presence in many bio-fluids, including those that can be collected with minimum harm to the patient, is one of the main advantages. This pervasiveness despite the presence of RNases has prompted studies that further supported their stability against the enzymes, as they resisted degradation. Stability studies have been conducted in some studies, where the lncRNAs in question were exposed to freeze-thaw cycles or stored at various temperatures (from room to higher temperatures) to further validate their robustness as biomarkers. A study was performed on breast cancer patients to investigate the potential of HIT as a diagnostic biomarker in plasma. The results revealed a significant change in the levels of HIT between the cancer and control groups, with higher sensitivity and specificity for CAl53 and CEA. PVT1 is another lncRNA that was proven to be upregulated in cancerous tissues, such as gastric cancer, hepatocellular carcinoma, NSCLC, bladder cancer, thyroid cancer, and malignant pleural mesothelioma, compared to normal tissues. BLACAT1, a novel promising biomarker lncRNA, was found to be highly expressed in multiple human cancers, such as Small-Cell Lung Cancer (SCLC) tissues.
A meta-analysis study was conducted to explore the diagnostic efficacy of lncRNAs between lung cancer patients and controls. The results highlighted higher sensitivity and AUC levels for lncRNAs (0.82 with 95% CI 0.79 to 0.84 and 0.88 with 95% CI 0.85 to 0.91, respectively), which were higher than those of already used serum markers. For instance, CEA has a sensitivity of 49.6%, while CYFRA21-1 has a sensitivity of 61.9%. Furthermore, this study found higher sensitivity, specificity and AUC levels for paralleled lncRNAs rather than single lncRNAs, with 0.86 vs. 0.80, 0.88 vs. 0.78 and 0.93 vs. 0.86, respectively. One last observation by this study indicated that lncRNA diagnostic efficacy levels in tissues were significantly lower than those in serum and plasma, with an AUC of 0.87 for tissues vs. 0.90 for serum or plasma. Such observation encourages lncRNAs as significant prospective diagnostic markers with minimally invasive procedures.
One example of lncRNA that was approved by the US Food and Drug Administration (FDA) for clinical usage is Prostate Cancer Antigen 3 (PCA3). The Progensa PCA3 test kit is now available in private hospitals and clinics to aid in decision making regarding prostate cancer patients, and this kit depends mainly on urine samples, which were proven to have significant levels of this RNA.
Mesothelioma
Malignant Mesothelioma (MM) is a cancer of the mesothelium, cells lining the pleura, peritoneum, pericardium, and tunica vaginalis. According to the WHO, pleural mesothelioma MPM, which comprises approx. 80% of all cases, is classified into 3 histo-pathological subtypes: Epithelioid, biphasic, and sarcomatoid.
Epithelioid cells are oval, polygonal or cuboidal cells that can mimic normal mesothelial cells. Sarcomatoids are mainly spindle cells. Biphasic cells are composed of both sarcomatoid and epithelioid cells within the same tumor.
Epithelioid is the most common subtype among the three. The epithelioid and biphasic subtypes make up 75–95% of all cases of diagnosed MM. The epithelioid subtype has the most favorable prognosis, with a median survival of 13 months. On the other hand, a diagnosis of MPM with a sarcomatoid subtype has the least favorable prognosis, with a median survival of 4 months. MM is predominantly associated with prior exposure to a group of carcinogenic silicate fibers called asbestos. It can be found as 6 chemically and physically diverse varieties (chrysotile, crocidolite, amosite, anthophyllite, tremolite, and actinolite), which may be generally subclassified according to the morphology of the fibers into 2 types: Serpentine (curved: Serpent-like) or amphibole (straight, needle-like).
Certain studies have suggested that amphibole asbestos fibers may have more risk than serpentine, likely due to their structure enabling easier inhalation. The global 5-year period prevalence of MM was 0.48 per 100,000 persons. As a result, it is considered a rare cancer. However, according to recent epidemiological investigations, the number of patients suffering from mesothelioma is on the rise, specifically in developing countries. Dramatic increases in cases have been observed at an estimated burden of 38,400 cases per year worldwide, despite international pushes to ban asbestos, with a formal ban enforced in 70 countries based on the lists of current asbestos bans and restrictions by the international ban asbestos secretariat, revised as of July 15, 2019.
The cause has been attributed to continued poor regulation of asbestos mining in countries such as India, Brazil, and Russia along with its general abundance in industrial and household usage.
MM is predominantly associated with prior exposure to a group of carcinogenic silicate fibers called asbestos. The development of the disease is latent, arising 30 to 40 years after the first exposure. This unusual latency period might explain the fact that the majority of MPM patients are 60 years old and older.
Other risk factors are indirect (para) exposure via a partner or relative working in occupations with asbestos exposure, as mentioned above. Environmental exposure could also become a risk factor for developing diseases, such as living near an asbestos factory. Rare heritable cases were linked to a mutation of the BAP1 gene.
In MPM patients, due to the prolonged onset of disease and complex nature of the disease, a definitive differential diagnosis is often difficult to make. Once one is established, treatment is discussed based upon the condition and progression of the cancer in the patient, with earlier diagnoses projecting better outcomes.
Patients who present with troubles in breathing or shortness in breath (dyspnea), chest pains and weight loss in addition to environmental or occupational exposure to asbestos are suspected of having MM. However, even with confirmed previous exposure, clinical manifestations are not enough to confirm the diagnosis alone since they are nonspecific. Thus, further tests are performed to reach a differential diagnosis. Physical performance is performed, and unilateral effusions are commonly observed.
Another option would be the use of imaging tests, which may give an indication for the presence of MPM, with the most sensitive imaging modality for diagnosis, evaluation and follow-up being a chest CT scan. CT scans cannot be used alone for a differential diagnosis and require more diagnostic tools for a final definitive diagnosis. Chest Radiography (CXR) is performed early on in the investigation; however, it has been shown to exhibit low sensitivity. Significant pleural effusions may mask pleural lesions and hide small pleural lesions caused by malignant cells (MPE-Malignant Pleural Effusion). Moreover, they lack the ability to detect the disease early. While biopsies are performed and aid tremendously in diagnosis, MPM is known to seed biopsy sites with tumor sites, which is why biopsies must be performed in the least invasive way. Even so, cases with extensive disease may make a thoracoscopy or open pleural biopsy more challenging. One method of establishing a diagnosis of MPM when a biopsy is not possible is to evaluate cytology specimens obtained from pleural effusions through thoracentesis. However, they are reported to have a level of variability and are thus not recommended by some guidelines.
To a lesser degree, immunohistochemistry may also be used as an adjunct. In fact, immunohistochemical staining of MPM-specific marker proteins such as calretinin, keratins 5/6, Wilms Tumor protein 1 (WT-1), and podoplanin are used as adjunct tools for diagnosis confirmation as well as to differentiate MPM from other tumor types. The accuracy of diagnosis can be further approved through the coupling of measuring IHC markers with a pleural biopsy. Furthermore, several of these markers were evaluated for their prognostic factors, such as calretinin, a known IHC marker for mesothelioma, which was evaluated by as a prognostic marker to measure whether patients would benefit from Extrapleural Pneumonectomy (EPP) as a therapeutic intervention.
However, few of the markers established have been shown to have high enough specificity and sensitivity. Furthermore, the selection of the marker depends on many factors, such as the sex of the patient, clinical findings, location and histologic features, as well as which would provide the best quality staining in a given laboratory.
LncRNAs as biomarkers for mesothelioma
Several novel biomarkers have been evaluated for mesothelioma, mostly as diagnostic markers. Many avenues have been investigated as potential biomarkers for diseases, most recently mRNA, miRNA, and antibody targets. One of these growing avenues is the untapped potential in the use of long noncoding RNA as biomarkers. There are various methods that have already been established, such as imaging screening, bronchoscopy, autoantibodies and liquid biopsies. Each of those techniques has its own limitations, whether due to their high costs, low sensitivity, over diagnosis or invasiveness. Researchers are endeavoring to uncover new, fast, cheap, relatively noninvasive and easy diagnostic tools. One such process was directed toward tumor biomarkers that are obtained from noninvasive samples such as urine, saliva, plasma, serum, stools, sputum and others.
Several novel biomarkers have been evaluated for mesothelioma, many as diagnostic or screening markers specifically. Many avenues have been investigated as potential biomarkers for diseases, most recently mRNA, miRNA, and antibody targets. One of these growing avenues is the untapped potential in the use of lncRNAs as biomarkers.
Aberrant lncRNA expression has been observed and validated in many cancers/tumors, which points to potential roles that have not yet been completely understood in cancer biology. One possible and often explored method of diagnosis is measuring biomarkers in bodily fluids. NcRNAs have been found in fluids such as serum/plasma that require minimal invasiveness to collect them, especially when compared with tissue biopsy or cerebrospinal sampling, which are more difficult and often carry some risk. Circulatory biomarkers in general are noninvasive, low-cost, and fast and require low technical expertise. Additionally, they have been found to be stable, with possibly higher diagnostic accuracy than conventional markers.
Taking all these points into account, the aim of this study was to explore whether lncRNAs could serve as appropriate circulatory biomarkers for the diagnosis of MPM. A systematic review was conducted to establish whether there was potential in exploring their potential further, and the results confirm that there is much yet to be explored and further lends support to the aim.
Literature Review
The overall framework for review
This review was performed according to the Preferred Reporting Items for systematic reviews and meta-analysis checklist (PRIMSA-P, 2015) and STAR-D in conjunction with Arksey and O’Malley’s six main framework stages:
• Identifying the research question (Using the PICO method (82)).
• Identifying relevant studies.
• Study selection.
• Charting the data.
• Collating, summarizing, and reporting the results.
• Consultation (optional and thus omitted).
Search process and literature selection
Search methods: Five databases were searched using the combination of “mesothelioma”, “lncrna” or “long noncoding RNA”. The searched databases were PubMed, JStor, MDPI, Scopus, and Science direct.
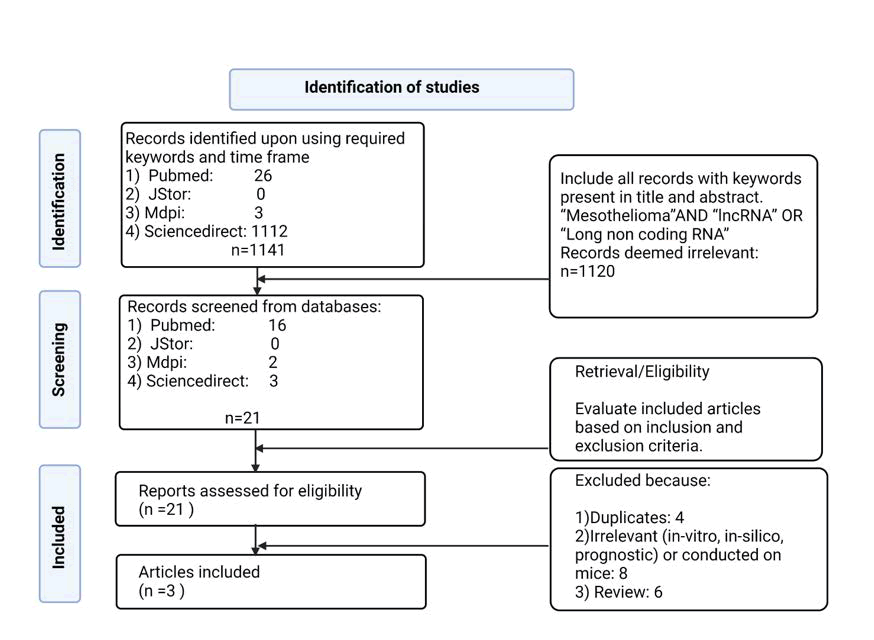
Records were from 2010 to 2022 to be based on more recent literature, and initial relevance was screened by title. Titles clearly referring to mesothelioma and long noncoding RNAs were further evaluated, and their abstracts were skimmed to evaluate their relevance to the topic (Figure 1).
Figure 1: PRISMA flow diagram for new systematic reviews, which included the study identification, screening and selection process.
Study eligibility
Inclusion criteria: Eligible studies met the following criteria:
• Patients in the study were diagnosed with malignant mesothelioma (MM).
• The study investigated the relationship between lncRNA expression levels and the diagnosis of MM.
• The study was published in English.
Exclusion criteria: Studies were excluded due to one or more of the following:
• Duplicate studies.
• Narrative review articles, case reports, and conference or poster abstracts.
• Expression levels of lncRNAs not measured by RTâ??qPCR ( by microarray or predictions).
• Studies conducted on animals.
• Insufficient data for extraction (for example, lack of sensitivity and specificity data).
Quality assessment: The quality of each included study was assessed by Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool.
Results
Characteristics of the included studies
The total number of studies screened was 433 (Figure 1). Twenty two were deemed relevant to the lncRNA and mesothelioma according to their titles and abstracts, and the full text was read to search for the inclusion and exclusion criteria. Four were removed due to being duplicates, 8 were excluded because the study was performed on mice or because it was irrelevant to the nature of the research question (for example, prognostic) or because the studies were in silico or microarray data, and 6 others were excluded due to being reviews. Subsequently, 3 articles were considered eligible, and their quality was evaluated according to QUADAS-2 (Table 2).
|
|
Weber et al. |
Matboli et al. |
Wright et al. |
|
Study design |
Case-control |
Case-control |
Cohort |
|
Country |
Berlin, Germany |
Cairo, Egypt |
Leicester, England |
|
Number |
22/44 |
60/40 |
14/3 |
|
Mean age (Patient/Control) |
60.1/55.2 |
71.5/72.5 |
60.1/55.2 |
|
Male/Female |
22/0 |
35/25 |
11/3 |
|
Histological subtype |
|||
|
Epithelioid |
14 |
57 |
5 |
|
Biphasic |
2 |
3 |
6 |
|
Sacromatoid |
2 |
0 |
3 |
|
Unknown |
4 |
0 |
1 |
|
Smoking status |
|||
|
Ever |
9 |
33 |
N.A |
|
Never |
11 |
27 |
N.A |
|
Unknown |
2 |
0 |
N.A |
Table 2: Overview of the kinds of patients/samples selected for the review from the studies.
Overall completeness and applicability of the data from the included studies
Quality of the evidence: As stated in the methods, the quality of each included study was assessed by the Quality Assessment of Diagnostic Accuracy Studies (QUADAS)-2 tool by 2 independent researchers. The results were generally good, and all studies scored 7 or higher in the assessment.
Three studies were included to evaluate the potential use of lncRNAs in the diagnosis of mesothelioma only. The sample size amounts to a total of 96 MM patients from various regions.
Potential biases in the review process: We adhered to the PRISMA checklist and established a search strategy that was followed by 2 researchers independently.
A possible source of bias is the exclusion of studies not published in English.
Data extraction: Data extracted from the articles include first author, publication year, nature of sample source, name of lncRNAs, and patient-control demographics.
Level of sensitivity, specificity, and AUC (ROC–Receiver Operating Characteristic) value (area under the ROC), and the expression of lncRNAs were collected as well as relevant information including but not limited to the sample size, reference gene, normalization method, and literature reference.
LncRNA as biomarkers for MM
Different types of biomarkers value shown in Table 3.
| LncRNA | Region | n | Source | Expression | Sensitivity | Specificity | AUC | Normalization | Quantification method |
| GAS5 | West Europe | 22 | Plasma | Up | 14 | 97 | 0.86 | RPLP0 | 2-ΔΔCt |
| lncRNA-RP1-86D1.3 | North Africa | 60 | Serum | Up | 83.3 | 95 | 0.876 | Hs_ACTB_1_SG QuantiTect Primer Assay (NM_001101) | 2-ΔΔCt |
| SNHG8 (NR_003584) | West Europe | 14 | MPM tissue | Up | 78.5 | 100 | 0.905 | 18S rRNA | 2-ΔΔCq |
| POT1-AS1 (BX648695) | West Europe | 14 | MPM tissue | Up | 78.6 | 100 | 0.93 | 18S rRNA | 2-ΔΔCq |
Table 3: Summary of results
Discussion
Upon examination of the literature, limited studies exist on the potential use of long noncoding RNAs as diagnostic biomarkers for malignant mesothelioma. The use of lncRNAs clinically has been a novel avenue that has yet to be explored, and consequently, a small number of studies have been found, especially with rare cancers such as mesothelioma, which further limits the number of studies performed on the topic. An overview of each lncRNA found through the systematic search is highlighted below.
GAS5
Growth arrest-specific 5, in which its gene locus is 1q25.1, was originally isolated by a subtractive cDNA library from mouse NIH 3T3 cells. Human GAS5 contains multiple C/D box small nucleolar RNAs (snoRNAs/SNORDs) coded within its 10 introns and 13 exons that can undergo alternative splicing to form at least 29 different mature RNA isoforms, 2 of which are long noncoding RNAs. The 2 mature lncRNA isoforms (reference sequences NR_002578.2 and AF141346.1) originate from the alternative splicing of 5’ donor sites at exon 7.
GAS5 is a member of the (5’TOP) gene family, as demonstrated by mapping of its 5’ end and the display of the characteristic unusual oligopyrimidine terminal sequence. This class of genes is known to accumulate in mRNA particles during arrested cell growth.
GAS5 has been found to have a tumor suppressor role phenotype, whereas dysregulation of its expression levels was found in many cancers, implying a role in their pathogenesis.
GAS5 was found to be down-regulated in several MPM cell lines. In a study by Renganathan et al., the role of GAS5 in the pathogenesis of MPM was evaluated in primary MPM cultures derived from surgical specimens. Expression levels were measured by RT-PCR, and GAS5 was found to be down-regulated in several MPM cell lines compared to normal mesothelial cells. On the other hand, intra tumoral levels obtained from the patients were higher than those in normal mesothelial tissues, whereas GAS5 levels are typically found to be down-regulated in other cancers 78 79.
However, in one study, GAS5 was found to be up-regulated significantly in both mesothelioma cell lines and the plasma of patients after an initial in silico analysis of published expression data derived from tissue samples of pleural mesothelioma.
Weber et al. measured GAS5 expression in 4 mesothelioma cell lines (NCI-H28, NCI-H2452, JL-1, and MSTO-211H), with fold changes greater than 2.0 representing upregulation and fold changes less than 1.5 representing down-regulation. Gas5 was found to be upregulated in 3 of the 4 cell lines used (at fold changes of 4.08, 8.52, and 2.64, with 1.19-fold being excluded according to the predetermined cutoff), which prompted further investigation of the lncRNA in the plasma levels of 22 cancer patients versus 44 asbestos exposed controls within the same study to validate the in vitro findings.
GAS5 was found to improve the performance of the panel as a third marker to calretinin and mesothelin at a fixed specificity of 97%, and the sensitivity increased to 73% with the linear combination and 68% with the sequential combination. The two-marker panel alone had a sensitivity of 46% at a predefined specificity of 98% in a separate study.
Differences in expressions between the studies could be attributed to different causes. LncRNAs are known to be tissue and cell specific, and they may exhibit differential expression during different pathologies and at different stages of the same diseases due to alternative splicing.
LncRNAs are almost universally alternatively spliced. Moreover, the spliced transcripts may display entirely different functions from each other, as was the case with LINC00477, a lncRNA spliced into 3 different transcripts in gastric cancer. Isoforms 1 (L1) and 2 (L2) were opposing each other both in expression levels and their role in the pathogenesis and progression of the cancer, as 1 was downregulated while 2 was upregulated when compared to normal cells. Similar variations in transcript expression have been observed in other cancers.
Interestingly, lncRNAs have also been observed to be regulators of alternative splicing.
While alternative splicing hints at a large diversity of variants, they may still have the same function and likely expression levels as well, provided they retain the same functional site postsplicing. Ultimately, the way to overcome this point of variation practically would be to design or choose a primer assay that detects as many spliced transcripts as possible or one that detects the most abundant isoform expressed in the disease and source of lncRNA if it has been previously reported.
Zhang et al., demonstrated that Gas5 directly targets miR-103 to reduce the phosphorylation of downstream proteins and hence inactivate the Akt/mTOR signaling pathway, with its overexpression inhibiting cell proliferation and cancer progression in prostate cancer. The Akt/mTOR pathway is commonly dysregulated in many cancers, including mesothelioma. Gas5 has been proven to display decoy functions by sequestering microRNAs in various other cancers.
The lncRNA has been shown to act as a regulator through this mode with miR-21, a microRNA that inhibits tumor suppressors PTEN and PDCD4. Consequently, this would overactivate the same Akt pathway mentioned above, as PTEN is reported to be a negative regulator through the miR-21/PTEN/Akt axis. This indicates that GAS5 inhibits this oncogenic pathway through several levels rather than one and adds further dimensions to the way lncRNAs work as regulators.
GAS5 has been consistently observed as a regulator of PTEN expression in other cancers through sponging of microRNAs.
GAS5 may also target proteins, as is the case with glucocorticoid receptors, as it acts as a decoy Glucocorticoid Response Element (GRE), thereby suppressing the induction of glucocorticoid-mediated transcriptional activity, a mechanism that may also be implicated in the pathology of mesothelioma, as high levels were found to suppress GR responsive genes that are involved in the cell cycle.
Other modes of action have also been reported, such as being a guide RNA for transcription factors and signal RNA, indicating the diverse nature of ncRNAs in performing their functions.
POT1-AS1
BX648695 or POT1-AS1 is one of the long noncoding RNAs, and its gene location is 7q31.33. This lncRNA was investigated in MPM tissues from EPP patients against cryopreserved benign pleural tissues. RT-PCR was used in this investigation and revealed significant differences in the expression level of POT1-AS1, with up-regulation in MPM compared to benign pleura by 2.95-fold. The ability of POT1-AS1 to differentiate between both was explored by Receiver Operating Characteristic (ROC) curve analysis, which showed that it has 93% accuracy, 78.6% sensitivity and 100% specificity. The AUC was 0.93 (95% confidence interval between 0.793-1.064, p=0.023) (Table 2).
Very few studies have highlighted BX648695 levels in cancer, such as a study performed on gastric cancer tissue samples. RT-PCR was used in this study and revealed upregulated levels in gastric cancer tissues compared to normal counterparts. The higher the levels of BX648695, the shorter the disease-free survival, as well as lower overall survival rates.
In view of this fact, BX648695 is considered an oncogenic lncRNA that needs extensive investigation, especially in the serum of MPM patients, for potential use as a diagnostic marker for these patients.
However, the underlying mechanism by which POT1-AS1 contributes to oncogenesis requires further investigation.
SNHG8
NR_003584, also known as lncRNA-small nucleolar RNA host gene 8 (SNHG8), has a gene locus at 4q26. It was investigated in the serum of breast cancer patients after collecting blood samples from two patients and one healthy patient. Exosomal RNA was extracted, cDNA synthesis was completed, amplification and purification by PCR were carried out, sequencing was performed, and the data were compared with known gene sequences revealing the downregulation of SNHG8 in comparison to healthy individuals.
Moreover, SNHG8 levels were also detected in tissues of other cancers, such as ovarian cancer, where it was overexpressed in comparison to healthy counterparts. Higher SNH8 levels were correlated with poorer prognosis, as well as enhancement of cancer invasiveness and proliferative abilities.
Similarly, when the levels of SNHG8 were investigated in prostate cancer tissues, it was found to be up-regulated as well. These higher levels played a role in enriching the cancer’s proliferative and migratory abilities as well. Other studies were conducted on different cancerous tissues and proven to have up-regulated levels of SNHG8, for instance, gastric cancer tissues, non-small cell lung cancer, colorectal cancer tissues and others.
Furthermore, it was one of the long noncoding RNAs that was investigated in MPM tissues from EPP patients along with cryopreserved benign pleural tissues. RT-PCR was used in this investigation and revealed up-regulated levels of SNHG8 in MPM compared with benign pleura by 5-fold. This was followed by an ROC curve that demonstrated the ability of SNHG8 to differentiate between benign pleura and MPM with a high degree of accuracy that reached 90.5%, 78.5% sensitivity and 100% specificity. The AUC was 0.905, the 95% confidence interval was 0.752-1.057, and P=0.33 (Table 2). Consequently, this can indicate the ability of this lncRNA to act as a diagnostic marker for MPM.
This differential expression of SNHG8 suggests that it has a great impact on cancer at both the prognostic and therapeutic levels. However, serum levels of SNHG8 were investigated in only one study to the best of our knowledge and on a very small number of patients, thus highlighting an interesting area of research to be inspected more in depth, especially in mesothelioma patients, as a potential diagnostic novel marker.
A study on non-small cell lung cancer identified miR-542-3p as a negatively regulated target of SNHG8. The study shows that overexpression of miR-542-3p leads to cell cycle arrest in G0/G1 phase through the inhibition of CCND1 and CDK6 expression. However, the detailed mechanism by which SNHG8-mediated gene expression participates in the progression of NSCLC remains unclear.
LncRNA RP1-86D1.3
LncRNA-RP1-86D1.3 or Long Intergenic Non-protein Coding RNA 1527 (LINC01527) is RNA expressed from LINC01527 gene locus 1q21.3. The expression levels of the novel lncRNA were found to be dysregulated in the tissue of CRC (colorectal cancer) and cell lines when compared to normal tissue. Moreover, it was found to be differentially expressed in gastric cancer after a pancancer analysis of 12 types of human cancer.
Hence, based on these findings and a bioinformatics analysis that was conducted, lncRNA-RP1-86D1.3 was chosen as a marker in a proposed diagnostic serum panel by. Based on the InCeDB database, the lncRNA was found to be a ceRNA regulating DRAM mRNA and ARSA mRNA, and 2 other markers of the panel were found to have significant differential expression and tissue specificity through two major databases, namely, the gene atlas database and the UniProt database.
LncRNA-RP1-86D1.3 was found to be elevated in the serum of 100 mesothelioma patients derived from the Egyptian population. At an optimum cutoff point of 1.31 (sensitivity, 83.3%; specificity, 95%, AUC (SE)=0.876 (0.0318), 95% confidence interval=0.802-0.950; P<0.01) according to ROC curve analysis, lncRNA-RP1-86D1.3 could be used to discriminate between healthy individuals and MPM patients. When further combined with other potential markers, the panel reached 100% sensitivity, 85% specificity and 94% accuracy in diagnosing MPM.
However, in one study, in silico analysis was performed using microarray expression data of altered lncRNAs from public gene expression repositories. Forty lncRNAs were found to be differentially expressed in pleural mesothelioma and normal human pleura, and RP1-86D1.3 was not one of them. However, differences may be attributed to differences in microarrays, sources of samples and the number of samples analyzed from which the data are retrieved. A larger prospective study is needed to fully validate the results.
lncRNA-RP1-86D1.3 has been implicated in other malignancies as a dys-regulated element, but the nature of its role, if any, is still to be determined, as it is more novel and less researched than the other aforementioned lncRNAs.
Conclusion
Current approved diagnostic methods have limitations that support a strong need to identify a highly sensitive, highly specific, and noninvasive method for the detection of MM.
One possible method of diagnosis is measuring biomarkers in bodily fluids. Various molecular markers were evaluated and were found to have potential in the diagnosis of cancers, one such marker being novel lncRNAs.
Limitations of this Study
The number of studies included in this review was limited owing to the nature of the cancer and the novelty of long noncoding RNAs. Hence, a larger pool of research is required to further support this conclusion. Each lncRNA appeared in one study, with only 1 or 2 being investigated in more than one study. Hence, validation studies with a larger, more diverse pool of patients are needed to investigate the expression of various lncRNAs. Moreover, all the data in the 3 studies were based on patients with the pleural form of mesothelioma, which, while it is the most common form, limits the breadth of the conclusion and excludes other forms, such as peritoneal or pericardial mesothelioma.
A more uniform preparation methodology would need to be adapted at various stages, from blood preparation to qRTâ??PCR analysis. Moreover, there was a lack of transparency in how the sample size was calculated, which according to the STARD checklist is recommended to evaluate whether the outcome has enough power to be generalizable or adequate to the objectives outlines by the study.
Different types of samples were used as sources for the extraction. LncRNAs may act in a tissue-specific manner, and as a result, their expression levels may vary accordingly. LncRNAs extracted from tissues seemed to perform better as biomarkers than circulating lncRNAs. However, more uniform protocols would need to be investigated and validated to enable a more homogenous comparison, as each study used different kits, gene expression assays/primers, or housekeeping genes.
Although all studies used RTâ??qPCR as their standard method of measuring lncRNA levels, the fold levels used to determine high/low levels were different in each study, introducing variation in the interpretation of results. For example, according, altered expression of lncRNAs was considered significant for fold changes <0.5 and >2.0. With, the lncRNA was included as overexpressed if the magnitude of change was >3-fold. Fold change cutoffs often vary according to the source of the lncRNA, with cells and tissues having more rigorous cutoffs than sources that usually contain low total RNA in general, such as serum or plasma.
The choice of primers may also affect the results. Ideally, primers should be able to detect all Expressed Sequence Tags (ESTs) of the lncRNA to detect all possible splice variants that may exist. However, some lncRNAs have a large number of isoforms, in which case the best coverage primers or assays should be chosen.
Furthermore, different reference genes were used for normalization, as there has not been a validation study to evaluate a specific reference gene suited for studying long noncoding RNA. Further studies have shown that certain housekeeping genes become inconsistent and fluctuate due to biological or pathological reasons, which cause inaccuracy in the relative quantification of lncRNAs using the prevalently used ΔΔCt method.
Common reference genes include GAPDH, ACTB, and small nuclear RNA U6 used for normalization.
Certain studies have evaluated the suitability of reference genes for specific cancers, such as the findings of Lempridee et al, whose results showed that lncRNAs RP11-204K16.1, XLOC_012542, and small RNA U6 were the optimum reference genes for circulating lncRNA analysis in cervical cancer, as they were stably expressed and not affected by age or hemolysis.
Assessing hemolysis by spectrophotometry after in particular may be an important step, as certain studies have found that it may have an impact on the levels of certain microRNAs; moreover, peripheral cells have been known to contain lncRNAs. RBC rupture may occur during sample handling or processing, which creates a level of variability that may need to be accounted for.
In conclusion, although a limited number of studies have been found, the preliminary data derived from these studies seem to point toward a hidden potential for the utilization of lncRNAs as biomarkers for mesothelioma and reveal a novel avenue worth exploring further, as there still exists a need for more reliable and straightforward methods for the disease. However, more studies need to be done to fully validate their function while also taking into account any limitations encountered in the previous studies.
References
- The ENCODE Project Consortium (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816.
[Crossref] [Google Scholar] [PubMed]
- Mattick JS (2005) The functional genomics of noncoding RNA. Science 309: 1527-158.
[Crossref] [Google Scholar] [PubMed]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Nonâ?ÂÂÂcoding RNAs: Regulators of disease. J Pathol 220: 126-139.
[Crossref] [Google Scholar] [PubMed]
- Dozmorov MG, Giles CB, Koelsch KA, Wren JD (2013) Systematic classification of non-coding RNAs by epigenomic similarity. InBMC Bioinform 1-12.
[Crossref] [Google Scholar] [PubMed]
- Vencken SF, Greene CM, McKiernan PJ (2015) Non-coding RNA as lung disease biomarkers. Thorax 70: 501-503.
[Crossref] [Google Scholar] [PubMed]
- Quinn JJ, Chang HY (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47-62.
[Crossref] [Google Scholar] [PubMed]
- Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, et al. (2019) LNCipedia 5: Towards a reference set of human long non-coding RNAs. Nucleic Acids Res 47: D135-D139.
[Crossref] [Google Scholar] [PubMed]
- The ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74.
- Clark DP, Pazdernik NJ, McGehee MR (2019) Chapter 19-noncoding RNA. Mol Biol 2019:604-621.
- Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22: 96-118.
[Crossref] [Google Scholar] [PubMed]
- Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10: 924-933.
[Google Scholar] [PubMed]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155-159.
[Crossref] [Google Scholar] [PubMed]
- Wei JW, Huang K, Yang C, Kang CS (2017) Non-coding RNAs as regulators in epigenetics. Oncol Rep 37: 3-9.
[Crossref] [Google Scholar] [PubMed]
- Dykes IM, Emanueli C (2017) Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics 15: 177-186.
[Crossref] [Google Scholar] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M (2013) Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 425: 3723-3730.
[Crossref] [Google Scholar] [PubMed]
- Hirota Y, Lahti JM (2000) Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res 28: 917-924.
[Crossref] [Google Scholar] [PubMed]
- Inoue A, Hyle J, Lechner MS, Lahti JM (2011) Mammalian ChlR1 has a role in heterochromatin organization. Exp Cell Res 317: 2522-2535.
[Crossref] [Google Scholar] [PubMed]
- Shah N, Inoue A, Lee SW, Beishline K, Lahti JM, et al. (2013) Roles of ChlR1 DNA helicase in replication recovery from DNA damage. Exp Cell Res 319: 2244-2253.
[Crossref] [Google Scholar] [PubMed]
- Grossi E, Raimondi I, Goni E, Gonzalez J, Marchese FP, et al. (2020) A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat Commun 11: 936.
[Crossref] [Google Scholar] [PubMed]
- Dai X, Kaushik AC, Zhang J (2019) The emerging role of major regulatory RNAs in cancer control. Front Oncol 9: 920.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi