Research Article, J Genet Disor Genet Rep Vol: 12 Issue: 6
Involvement of Precancerous Lesions Risk Factors in Epigenetic Peculiarity of Oncogenic Viruses of Alphapapillomavirus Genus Based on the CpG Dinucleotides Island Variability
Embolo Enyegue Elisee Libert1*, Ndipho Tatou Christian Kitchener1, Awalou Halidou1, Ananga Noah Sidonie2, Halmata Mohamadou1, Godwe Celestion3, Eyebe Richard Honore1, Evina Evina Basile4, Bell Eric Michel5 and Essame Oyono Jean Louis1
1Department of Medical Research, Institute of Medical Research and Studies on Medicinal Plants, Yaounde, Cameroon
2Department of Medicine and Pharmaceutical Sciences, University of Douala, Douala, Cameroon
3Department of Medicine, University of Washington, Seattle, Washington
4Department of Pharmaceutical Sciences, Catholic University of Louvain, Louvain, Belgium
5Department of Health Science, University of Bamenda, Bamenda, Cameroon
- *Corresponding Author:
- Embolo Enyegue Elisee Libert
Department of Medical Research,
Institute of Medical Research and Studies on Medicinal Plants,
Yaounde,
Cameroon, 237671701458;
E-mail: eliseeembolo@yahoo.fr
Received date: 05 May, 2023, Manuscript No. JGDGR-23-97914;
Editor assigned date: 08 May, 2023, PreQC No. JGDGR-23-97914 (PQ);
Reviewed date: 22 May, 2023, QC No. JGDGR-23-97914;
Revised date: 05 July, 2023, Manuscript No. JGDGR-23-97914 (R);
Published date: 12 July, 2023, DOI: 10.4172/2327-5790.1000226
Citation: Libert NEEE, Kitchener NTC, Halidou A, Sidonie AN, Mohamadou H, et al. (2023) Involvement of Precancerous Lesions Risk Factors in Epigenetic Peculiarity of Oncogenic Viruses of Alphapapillomavirus Genus Based on the CpG Dinucleotides Island Variability. J Genet Disor Genet Rep 12:6.
Abstract
Background: Cervical lesions are changes in cervical cells that make them more likely to develop into cancer. Many risk factors are involved in the occurrence of cancer; they may then alter the methylation profile of known oncogenic HPVs which is a ubiquitous epigenetic regulatory signature for any entity with DNA that occurs in CpG Islands. In order at appraising the CpG Islands profile for distribution on papillomaviruses genome.
Methods: An analysis of dinucleotides frequencies was carried out from DNAs of diverses Alphapapillomavirus genus. We focused our attention on 63 genomes of Alphapapillomavirus genus, looking for internal CpG Island profile. Bioinformatics software online dbcat and graph pad prism 8.0.1 for statistical analysis were used.
Results: The distribution of CpG Islands across HPV genes was not random; oncogenes had CpG Island in 100% of cases.
Conclusion: These findings demonstrate that each HPV genotype can have the potential to transform into a high risk genotype by losing one or more of its CpG Islands, and that this process may be accelerated by the presence of host related risk factors. Analysis of CpG Islands suggests that risk factors for the development of precancerous lesions could be involved in the HPV epigenetic profile change.
Keywords: Epigenetic; Alphapapillomavirus; CpG; Methylation; Oncogenic viruses
Introduction
Human Papillomaviruses (HPVs) are a long lived and highly effective group of viruses that have co-evolved with their hosts to replicate in certain stratified epithelia anatomical niches, to date, the genomes of almost 450 distinct HPV types have been isolated and sequenced. The viral replication cycle in these viruses is influenced by the differentiation of the infected epithelium. The viral replication cycle in these viruses is influenced by the differentiation of the infected epithelium [1]. The epidemiology of HPV infections reflects this genetic diversity of HPVs, with a specific geographical distribution of different variants of an HPV type. The genetic variety of HPVs may have an impact on the pathophysiology of HPV infection, it is known that co-infection with multiple types of HPVhr is a risk factor for cancer advancement and polymorphisms are linked to an increased risk of cancer progression. Determining the human papillomavirus’s potential methylation profile to help understand the purpose of this diversity remains a difficulty. Indeed, investigations have demonstrated that the virus’s DNA methylation is linked to how well it responds to treatment for the pathology it causes. Increased methylation was detected at 13 CpG sites with disease progression, and a high level of methylation was associated with the risk of CIN2+ in a study to determine the correlation between CpG methylation in Human Papillomavirus (HPV)-16 L1 and persistent infections and the development of cervical carcinoma in Uyghur women. These findings suggest that methylation of CpG sites in HPV could be useful in predicting HPV infection persistence and cervical pre-cancer development [2].
Materials and Methods
Type of study
This was a retrospective descriptive study.
Sample collection strategy
The sequences for analysis were taken from pave.niaid.nih.gov (Alphapapillomavirus). A total of 63 HPV genotypes sequences were extracted from pave.niaid.nih.gov a specialized HPV database [3].
Selection criteria for CpG Island selection
During the study, any region with a concentration of at least 200 pb, a GC concentration of at least 50%, and a CpG ratio of at least 60% were considered to be CpG Island.
Bio-informatical analyses
Sequence exploration was done using dbcat.cgm.ntu.edu.tw, which is an online methylation analytical tool, composed of three parts: A CpG Island finder, a genome query browser and an analytical tool for methylation microarray data. The analytical tool can analyze raw data generated from scanners and search genes with methylated regions which could affect gene expression regulation [4].
Statistical analysis
Statistical analyses were performed using graphpad prism 8.0.1.
Results
Distribution of the number of CpG Islands
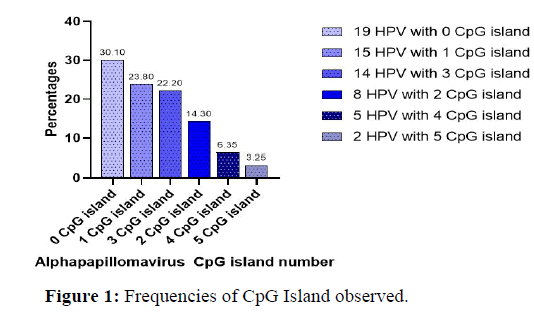
The distribution of the number of CpG Islands in all of the genomes analyzed is shown in Figure 1 below [5-9]. Indeed, many of the viruses we encountered lacked CpG Islands (30.1%). The viruses with the most CpG Islands made up only 3.25% of the total number of viruses in the research.
Profile of HPV without CpG Islands
The strains listed in Table 1 below were found to have no CpG Islands when their genomes were examined. Mucosal tropic viruses were found in all of these genotypes. The viruses without CpG Islands detected belonged to five Alpha p apillomavirus species (alpha HPV 6, alpha HPV 7, alpha HPV 9, alpha HPV 10, alpha HPV 11). There are no significant types in the alpha HPV 10 species these species are generally regarded a low risk virus that is commonly found in condyloma acuminate and laryngeal papillomatosis [10]. There are no major CpG Islands in the genomes of subtypes HPV 13, HPV 44, and HPV 73. We found HPV type 16 and 6 subtypes in Alpha papillomavirus 9, which contains the highrisk genotypes responsible for mucosal lesions [11]. These included HPV 31, HPV 33, HPV 35, HPV 52, HPV 58, and HPV 67. HPV 18, which included six subtypes, including HPV 39, HPV 45, HPV 56, HPV 68, HPV 70, and HPV 97, manifested as Alpha papillomavirus 7, which also has the high risk genotypes often responsible for mucosal lesions. Alpha papillomavirus 11 species and its major type, HPV 34, were also found, as well as Alpha papillomavirus 6 species and its major type, HPV 53 [12].
| CpG Island number | Species | Types species | Other Papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 0 | 10 | - | HPV 13 (X62843) | 19 |
| HPV 44 (U31788) | ||||
| HPV 73 (X94165) | ||||
| 9 | HPV 16 (K02718) | HPV 31 (J04353) | ||
| HPV 33 (M12732) | ||||
| HPV 35 (X74476) | ||||
| HPV 52 (X74481) | ||||
| HPV 58 (D90400) | ||||
| HPV 67 (D21208) | ||||
| 7 | HPV 18 (X05015) | HPV 39 (M62849) | ||
| HPV 45 (X74479) | ||||
| HPV 56 (X74483) | ||||
| HPV 68 (X67161) | ||||
| HPV 70 (U21941) | ||||
| HPV 97 (DQ080080) | ||||
| 11 | HPV 34 (X74476) | - | ||
| 6 | HPV 53 (X74482) | - | ||
| Total | 5 | 4 | 15 |
Table 1: Presentation of HPV with 0 CpG Island.
Distribution of genotypes with no CpG Islands according to their oncogenic potential
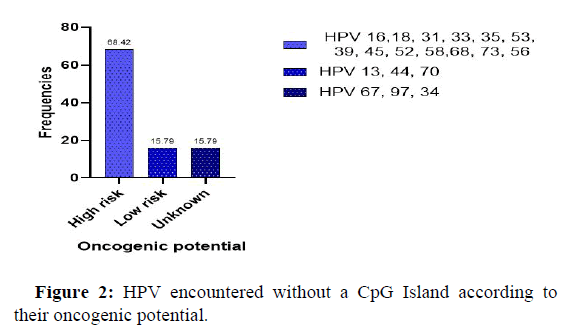
Figure 2 below shows the viral genotypes of HPV according to their oncogenic potential, which did not show any CpG Islands. It can be seen that high risk viruses represent about 69% of the viruses without CpG Islands in their genome, low risk oncogenic viruses are also observed with a frequency of 15.79%. However, we also observed viruses whose oncogenic risk is not really fixed that showed an absence of CpG Islands, notably viruses [13].
Distribution of CpG Islands in HPV genome
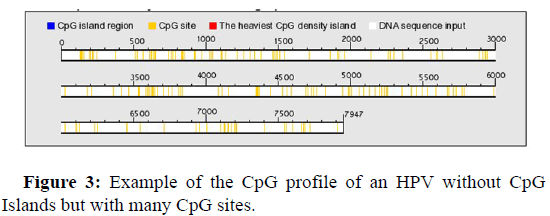
Figure 3 below, shows an example of the genomic profile of an HPV without CpG Islands, however we observe many CpG sites, which are not Islands and can probably also benefit from various methylations.
Profile of HPV with a single CpG Island
Table 2 below shows the different types of viruses encountered that had only one CpG Island. Indeed, 7 different species of Alpha Papillomaviruses were found which had 5 major and 10 minor papillomavirus types [14].
| CpG Island number | Species | Types species | Other Papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 1 | 10 | HPV 6 (X00203) | HPV 11 (M14119) | 15 |
| 8 | HPV 7 (X74463) | HPV 43 (AJ620205) | ||
| HPV 91 (AF419318) | ||||
| 5 | HPV 26 (X74472) | HPV 51 (M62877) | ||
| HPV 69 (AB027020) | ||||
| HPV 82 (AB027021) | ||||
| 1 | HPV 32 (X74475) | - | ||
| 6 | - | HPV 30 (X74474) | ||
| HPV 66 (U31794) | ||||
| 13 | HPV 54 (U37488) | - | ||
| 7 | - | HPV 59 (X77858) | ||
| HPV (KR816168) | ||||
| Total | 7 | 5 | 10 |
Table 2: Presentation of HPV with 1 CpG Island.
Genomic regions hosting the observed single CpG Islands
Table 3 below shows the different portions of the genome on which a CpG Island was encountered. In total 15 viruses had a CpG Island, of these genotypes 46.66% were high risk and low risk [15]. One HPV 177 virus had no oncogenic information, it had an unknown status. The vast majority of the CpG Island was identified in the E2 genes in almost 80% of the cases. The size of the CpG clusters ranged from 224 to 410 nucleotides.
| Sequences | Oncogenic potential | Sequence of each CpG Island | CpG Island length | Gene location |
|---|---|---|---|---|
| HPV6REF | LR | 3437-3710 | 274 | E2 |
| HPV7REF | LR | 3389-3734 | 346 | E2 |
| HPV11REF | LR | 3403-3714 | 312 | E2 |
| HPV26REF | HR | 3331-3684 | 354 | E2 |
| HPV30REF | HR | 842-1068 | 227 | E6-E7-E1 |
| HPV32REF | LR | 4545-4954 | 410 | L2 |
| HPV43REF | LR | 1267-1530 | 264 | E1 |
| HPV51REF | HR | 3399-3624 | 226 | E2 |
| HPV54REF | LR | 3307-3574 | 268 | E2 |
| HPV59REF | HR | 3433-3739 | 307 | E2 |
| HPV66REF | HR | 3370-3662 | 293 | E2 |
| HPV69REF | HR | 3327-3632 | 306 | E2 |
| HPV82REF | HR | 3431-3712 | 282 | E2 |
| HPV91REF | LR | 3399-3796 | 398 | E2 |
| HPV177REF | U | 4170-4393 | 224 | E2 |
Note : LR: Low Risk; HR: High Risk; U: Unknown
Table 3: CpG Island location on HPV.
CpG Islands positions on HPV
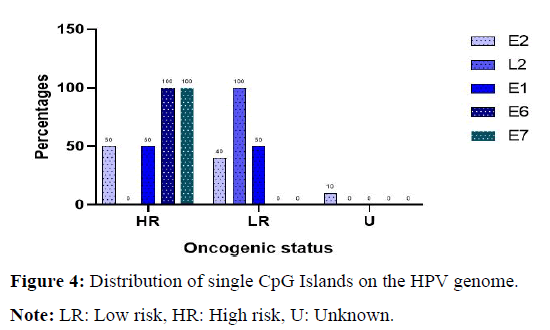
The Figure 4 below shows the distribution pattern of the Islands on the different Human papillomavirus genes. It can be seen that 50% of the CpG Islands are located on gene 2, for high risk genotypes, while for low risk genotypes, CpG Islands were found on gene E2 in 40% of cases. One genotype with unknown oncogenic potential also harbored CpG Islands on its E2 gene [16]. No CpG Islands were found in the L2 gene of the high risk genotypes, but the only Islands found in the L2 gene for viruses with a single Island were found in a low risk genotype. We were able to identify Islands at the E1 gene in equal proportions for both high and low risk genotypes. The HPV 30 genotype, which is a high risk genotype, showed that the CpG Island encountered overlapped 3 genes (E6-E7-E1).
Note: LR: Low risk, HR: High risk, U: Unknown.
Distribution of a CpG Island and CpG sites on the HPV genome
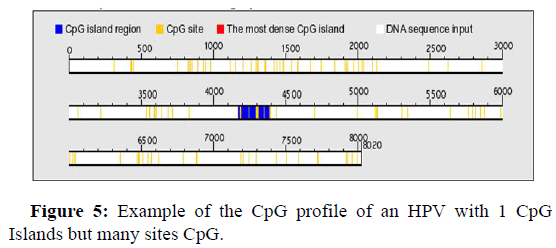
Figure 5 below, shows an example of the genomic profile of an HPV with single CpG Islands, however we observe many CpG sites, which are not Islands and can probably also benefit from various methylations.
Profile of HPV with two CpG Islands
The HPVs presented in the Table 4 below show a large absence of major HPV types. Alpha Papillomavirus species 2 showed no major types, compared to 3 minor types. HPV species 8, 6 and 3 showed only subtypes of HPV, while HPV species 15 showed a major type.
| CpG Island number | Species | Types species | Other Papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 2 | 2 | - | HPV 3 (X74462) | 8 |
| HPV 77 (Y15175) | ||||
| HPV 94 (AJ620211) | ||||
| 8 | - | HPV 40 (X74478) | ||
| 15 | HPV 71 (AB040456) | - | ||
| 6 | - | HPV 74 (U40822) | ||
| 3 | - | HPV 83 (AF151983) | ||
| 14 | - | HPV 106 (DQ080082) | ||
| TOTAL | 6 | 1 | 7 |
Table 4: HPV with two CpG Islands.
Genomic regions hosting 2 CpG Islands
Table 5 below shows the genomic areas of localisation on the viruses that showed 2 CpG Islands. Of these viruses, only one low risk virus was observed, most of the viruses encountered with this profile had no known oncogenic potential. The size range of the CpG Islands was between 1408 and 221 [17]. It was observed that about 50% of these CpG Islands were located at the E2 gene, 25% were found at the E1 gene, 18.75% of the CpG Islands were found at the L2, E6, E7 genes.
| Sequences | Oncogenic potential | Sequence of each CpG Island | CpG Island length | Gene location |
|---|---|---|---|---|
| HPV3REF | U | 3283-3809 | 527 | E2 |
| 4370-4661 | 292 | L2 | ||
| HPV40REF | LR | 1190-1518 | 329 | E1 |
| 3309-3748 | 440 | E2 | ||
| HPV71REF | U | 471-1456 | 986 | E6-E7-E1 |
| 3377-3795 | 419 | E2 | ||
| HPV74REF | U | 620-840 | 221 | E6 |
| 3199-3582 | 384 | E2 | ||
| HPV77REF | U | 3260-3880 | 621 | E2 |
| 4471-4703 | 233 | L2 | ||
| HPV83REF | U | 3231-3640 | 410 | E2 |
| 4439-5134 | 696 | L2 | ||
| HPV94REF | U | 85-1492 | 1408 | E6-E7-E1 |
| 3203-3808 | 606 | E2 | ||
| HPV106REF | U | 644-1491 | 848 | E7-E1 |
| 3188-3777 | 590 | E2 |
Note: LR: Low Risk; HR: High Risk; U: Unknown
Table 5: CpG Island location on HPV with 2 CpG Islands.
Position of CpG Islands on genes for viruses with 2 Islands
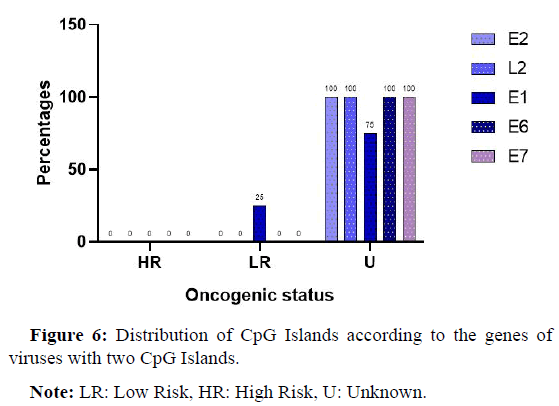
Figure 6 below shows the different identification sites of the various CpG Islands and the viral types in which they were found.
There was a complete absence of high risk viruses among the viruses that presented 2 CpG Islands, but one low-risk virus was present.
Almost all the viruses encountered had an unknown oncogenic potential and with almost 100% of the genes harboring CpG Islands apart from the E1 gene where we encountered CpG Islands at a frequency of 75%.
Note: LR: Low Risk, HR: High Risk, U: Unknown.
Distribution of two CpG Islands and CpG sites on the HPV genome
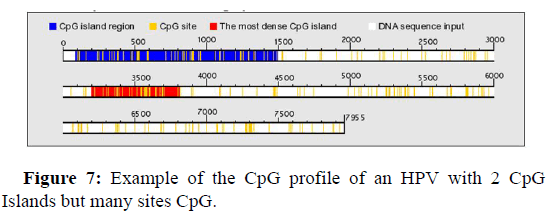
Figure 7 below shows an example of the distribution of CpG Islands in an HPV genome hosting 2 CpG Islands. A first CpG cluster is observed between nucleotides 100 and 1500 of the genome, however the densest region is observed between nucleotide 3200 and 3800.
Profile of HPV with three CpG Islands
Table 6 below shows the different Alphapapillomavirus species in which we were able to identify 3 CpG Islands. We observe that there were 5 species of Alphapapillomavirus that could be identified; only two species presented main HPV types (HPV 10 and HPV 14). The identified HPV subtypes were 12.
| CpG Island number | Species | Types species | Other Papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 3 | 2 | HPV 10 (X74465) | HPV 28 (U31783) | 14 |
| HPV 29 (U31784) | ||||
| HPV 78 (KC138720) | ||||
| HPV 117 (GQ246950) | ||||
| HPV 125 (FN547152) | ||||
| 4 | - | HPV 27 (X73373) | ||
| 1 | - | HPV 42 (M73236) | ||
| 3 | - | HPV 72 (X94164 | ||
| HPV 81 (AJ620209) | ||||
| HPV 84 (AF293960) | ||||
| HPV 89 (AF436128) | ||||
| HPV 102 (DQ080083) | ||||
| 14 | HPV 90 (AY057438) | - | ||
| Total | 5 | 2 | 12 |
Table 6: Presentation of HPV with 3 CpG Islands.
Genomic regions hosting 3 CpG Islands
Table 7 below shows the different viral genotypes that harboured 3 CpG Islands. In total 13 viral genotypes were found to contain three CpG Islands distributed in different viral genes. Among these viruses we identified 69.23% of viruses with unknown oncogenic potential against 30.76% of low risk viruses. The range of variation of the CpG Islands encountered was from 1470 to 233 nucleotides. No high-risk virus was observed among the viruses with 3 CpG Islands.
| Sequences | Oncogenic potential | Sequence of each CpG island | CpG Island length | Gene location |
|---|---|---|---|---|
| HPV10REF | U | 3377-3927 | 551 | E2 |
| 4514-4743 | 230 | L2 | ||
| 5100-5574 | 475 | L2 | ||
| HPV27REF | U | 436-1663 | 1228 | E6-E7-E1 |
| 3194-3813 | 620 | E2 | ||
| 5312-5598 | 287 | L2 | ||
| HPV28REF | U | 110-1579 | 1470 | E6-E7-E1 |
| 3248-3724 | 477 | E2 | ||
| 4422-4654 | 233 | L2 | ||
| HPV29REF | U | 3287-3916 | 630 | E2 |
| 4495-4879 | 385 | L2 | ||
| 5020-6175 | 1156 | L2-L1 | ||
| HPV42REF | LR | 1149-1468 | 320 | E1 |
| 3390- 3767 | 378 | E2 | ||
| 4506-4726 | 221 | L2 | ||
| HPV72REF | LR | 718-1603 | 886 | E6-E7-E1 |
| 3322-3754 | 433 | E2 | ||
| 4500-4986 | 486 | L2 | ||
| HPV78REF | U | 246-1526 | 1281 | E6-E7-E1 |
| 3252-3815 | 564 | E2 | ||
| 4422-4654 | 233 | L2 | ||
| HPV81REF | LR | 949-1509 | 561 | E7-E1 |
| 3438-3906 | 469 | E2 | ||
| 4553-4838 | 286 | L2 | ||
| HPV84REF | U | 480-1342 | 863 | E7-E1 |
| 3259-3741 | 483 | E2 | ||
| 6994-7216 | 223 | L1-URR | ||
| HPV89REF | LR | 65-1488 | 1424 | E6-E7-E1 |
| 3237-3673 | 437 | E2 | ||
| 4426-4649 | 224 | L2 | ||
| HPV102REF | U | 3270-3755 | 486 | E2 |
| 4460-4860 | 401 | L2 | ||
| 7540-7800 | 261 | L1-URR | ||
| HPV117REF | U | 328-1543 | 1216 | E6-E7-E1 |
| 3305-3871 | 567 | E2 | ||
| 4507-4739 | 233 | L2 | ||
| HPV125REF | U | 3173-3661 | 489 | E2 |
| 4291-4523 | 233 | L2 | ||
| 5538-6188 | 651 | L2-L1 |
Note: LR: Low Risk; HR: High Risk; U: Unknown
Table 7: CpG Island location on HPV with 3 CpG Islands.
Position of CpG Islands on genes for viruses with 3 Islands
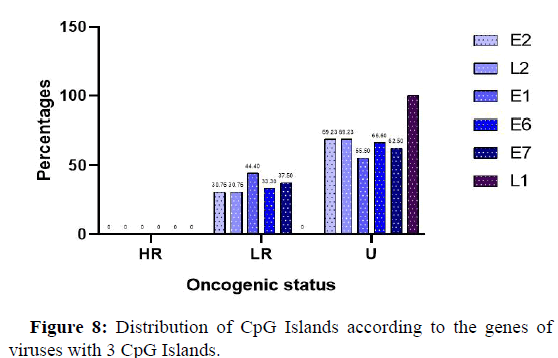
Figure 8 below shows the frequency of localization of the different CpGs on the viruses according to the genes. Indeed, it is observed that viruses with low oncogenic risk presented less than 45% of genes harboring Island CpGs, while genes with unknown oncogenic potential presented viruses that had more than 60% of the genes occupied by Island CpGs.
Distribution of three CpG Islands and CpG sites on the HPV genome
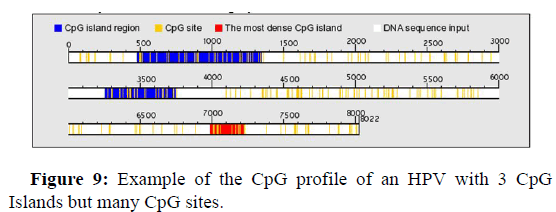
Figure 9 below shows an example of a viral genome with 3 CpG Islands. The first CpG Island can be observed between nucleotide 495 and nucleotide 1380, the second one Island can be observed between nucleotide 3250 and nucleotide 3750, while the densest Island is observed between nucleotide 7000 and nucleotide 7250.
Profile of HPV with four Islands
Table 8 below shows three species of HPV with one main type of HPV (HPV2) and three secondary HPV types.
| CpG Island number | Species | Types species | Other Papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 4 | 2 | - | HPV 160 (AB745694) | 4 |
| 4 | HPV 2 (X55964) | HPV 57 (X55965) | ||
| 3 | - | HPV 114 (GQ244463) | ||
| Total | 3 | 1 | 3 |
Table 8: Presentation of HPV with 4 CpG Islands.
Genomic regions hosting 4 CpG Islands
Table 9 below shows the different viruses on which 4 CpG Islands were found. It can be seen that all of these viruses (100%) were viruses of unknown oncogenic potency, the range of CpG Islands observed was between 1247 and 221.
| Sequences | Oncogenic potentiel | Sequence of each CpG island | CpG Island length | Gene location |
|---|---|---|---|---|
| HPV2REF | U | 542-1604 | 1063 | E7-E8 |
| 3218-3806 | 589 | E2 | ||
| 4451-4890 | 440 | L2 | ||
| 5067-5761 | 695 | L2-L1 | ||
| HPV57REF | U | 531-1660 | 1130 | E7-E1 |
| 2599-3777 | 1179 | E2 | ||
| 5012-6237 | 1126 | L2-L1 | ||
| 6420-6828 | 409 | L1 | ||
| HPV114REF | U | 909-1655 | 747 | E7-E1 |
| 3473-3964 | 492 | E2 | ||
| 4486-4905 | 420 | L2 | ||
| 7254-7474 | 221 | L1 | ||
| HPV160REF | U | 3379-3850 | 472 | E2 |
| 4569-4789 | 221 | L2 | ||
| 4982-6228 | 1247 | L2-L1 | ||
| 6414-6791 | 378 | L1 |
Table 9: CpG Island location on HPV with 4 CpG Islands.
Position of CpG Islands on genes for viruses with 4 Islands
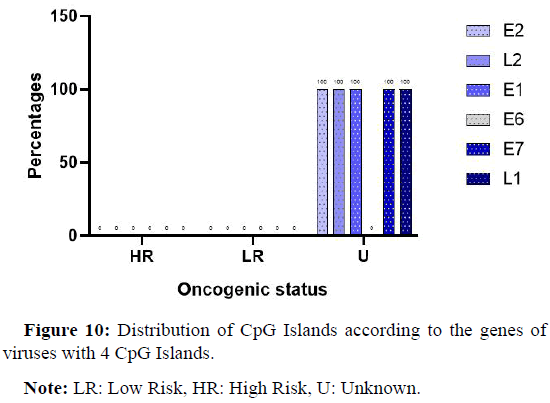
Figure 10 shows the distribution of CpG Islands on different genes. It can be seen that these Islands are 100% present on almost all the genes except for the E6 gene, where no CpG Islands were found.
Note: LR: Low Risk, HR: High Risk, U: Unknown.
Distribution of 4 Island CpG and CpG sites on the HPV genome
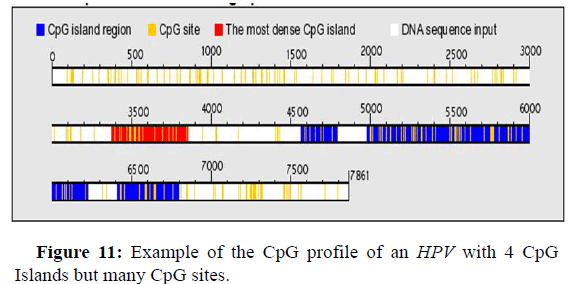
Figure 11 below shows an example of an HPV genome with 4 CpG Islands. The first very dense CpG Island is observed between Figure 11 below shows an example of an HPV genome with 4 CpG Islands. The first very dense CpG Island is observed between nucleotide 3400 and nucleotide 3850, a second Island is observed at nucleotide 4600 and nucleotide 4800, the third Island is observed at nucleotide 4990 and nucleotide 6220 and the last CpG Island observed is encountered between nucleotide 6400 and nucleotide 6800.
Profile of HPV with five CpG Islands
Table 10 below shows the HPV viruses in which 5 CpG Islands were observed. Indeed, 2 viruses among those selected showed 5 CpG Islands, they are the same species of alpha papillomavirus, species 3 which presented a main genotype (HPV 61) which has a low oncogenic risk and a secondary genotype (HPV 62) so the oncogenic power is unknown.
| CpG Island number | Species | Types species | Other papillomavirus type | Total number of HPV |
|---|---|---|---|---|
| 3 | HPV 61 (U31793) | HPV 62 (AY395706) | 2 | |
| Total | 1 | 1 | 1 |
Table 10: Presentation of HPV with 5 CpG Islands.
Genomic regions hosting 5 CpG Islands
Table 11 below shows the locations of the CpG islands encountered. The range of CpG islands is between 837 and 221.
| Sequences | CpG Island(s) found | Sequence of each CpG island | CpG Island length | Gene location |
|---|---|---|---|---|
| HPV61REF | LR | 802-1457 | 656 | E7-E1 |
| 3379-3742 | 364 | E2 | ||
| 4491-5025 | 535 | L2 | ||
| 5293-5584 | 292 | L2 | ||
| 7142-7362 | 221 | L1-URR | ||
| HPV62REF | U | 99-404 | 306 | E6 |
| 541-1377 | 837 | E7-E1 | ||
| 3316-3790 | 475 | E2 | ||
| 4463-4897 | 435 | L2 | ||
| 5102-5403 | 302 | L2 |
Table 11: CpG Island location on HPV with 5 CpG Islands.
Position of CpG Islands on genes for viruses with 5 Islands
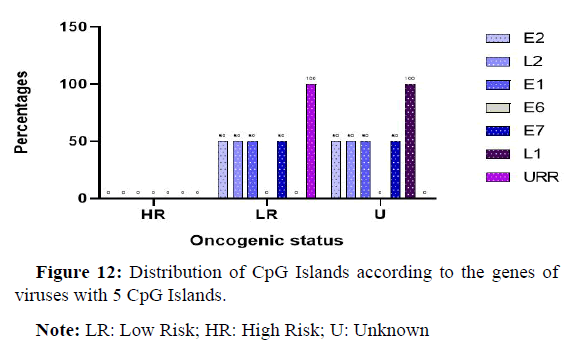
The Figure 12 below shows the distribution of the Islands detected on the different genes of the virus. Indeed, it can be seen that most of the genes harboured at least 50% of the CpG Islands, only the E6 gene which had no CpG Islands. The URR regulation regions had Islands in 100% of the cases.
Note: LR: Low Risk; HR: High Risk; U: Unknown
Distribution of 5 CpG Islands and CpG sites on the HPV genome
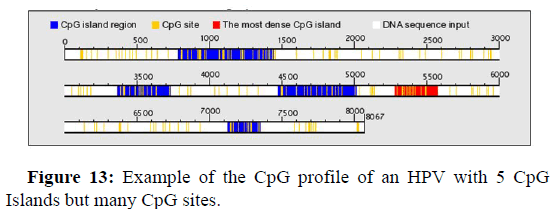
Figure 13 below shows an example of a virus genome hosting 5 CpG Islands. The first islet is located between nucleotide 800 and nucleotide 1420, a second islet is located between nucleotide 3370 and nucleotide 3710, the third nucleotide is located between 4480 and 5050, the fourth islet in this image, which is the densest islet, is located between nucleotide 5300 and nucleotide 5600, and the lastCpG islet in this image 13 is located between nucleotide 7120 and nucleotide 7350.
Discussion
This work was initiated in order to assess the distribution of CpG Islands profiles on papillomavirus genomes, and then to try to comprehend the impact of these predispositions on the neoplastic dynamics of different types of Alphapapillomavirus genus in the host cell. The preliminary results of this study show that, Human Papilloma Virus (HPV) genotypes show epigenomic variations, resulting in different CpG Islands in each HPV genotypes or sub genotype. This may be due to the fact that different human papillomavirus genotypes have different mucocutaneous tropisms, therefore, this could influence the position of CpGs within the genome these results are in agreement with the work of Chen and colleagues, who demonstrated a difference in the profile of CpG Islands in the hepatitis B virus. Some HPVs, however, did not show CpG Islands, this absence of CpG Islands in certain HPV viruses is seen in the majority of high risk HPV viruses, implying that the absence of the CpG Island in these viruses could be a crucial biomarker for their identification. It's also plausible that the absence of these CpG Islands was produced by the virus's oncoproteins altering the host environment. The work of XIA before and Thomas and Sudhir after had proposed the following the hypothesis that genomic methylation may be influenced by defenses specific to a host environment. According to their oncogenic potential, high risk viruses account for around 69% of viruses lacking CpG Islands in their genome, while low risk oncogenic viruses account for 15.79% of all viruses. This increased absence of CpG Islands in high risk viruses is a fact that could be a direct consequence of the evolution of these viruses as demonstrated in the study by Mohita, et al. who concluded that the extent of CpG depletion among papillomaviruses and polyomaviruses is linked to the evolutionary lineage of the infected host. Silvia and collaborator showed a positive correlation between CpG observed and expected values, with mucosal High Risk (HR) virus types showing the smallest O/E ratios. However, viruses with an undetermined carcinogenic risk, such as viruses, were found to be lacking in CpG Islands (HPV 67, HPV 97, and HPV 34). The fact that these viruses presented the same profile as the known oncogenic risk viruses would imply that these viruses would have an oncogenic potential within the host cells this observation is quite logical since genotypes 67 and 97 are subtypes of HPV 16 and HPV 18 viruses which have a known oncogenic potential, some authors isolated these genotypes from high grade intraepithelial lesions. In some viruses, only one CpG Island was found, this CpG Island was found in 46.66% of cases on high risk and low risk genotypes and 80% of the cases, the great majority of the CpG Island was found in the E2 genes. The presence of CpG Islands on E2 may be the regulatory role that this gene plays in viral replication. It is known that, viral E2 protein a sequence specific DNA binding protein regulates the papillomavirus replication cycle and can operate as a transcriptional activator or regulator of viral gene expression. Several other genes have CpG Islands, indicating the presence of methylation sites within these genes. Given that methylation is such an important phenomenon in the genome, the presence of these CpG Islands within these genes is tangible evidence of the virus's regulatory quality, although reversible. CpG Islands are important in the protection of adjacent housekeeping genes from de novo DNA methylation and for keeping them in a transcriptionally active state. In this work, we found that the size of the CpG Islands varied with the number of Islands encountered, large CpG Islands were more likely to be found in viruses that had several of them. Indeed, this aspect of things would probably be due to their oncogenic potential, which could be explained by the study by Yuan and colleagues, who demonstrated that the lengths and QS heterogeneity of CpG islets differ according to the clinical phases of an infection. These data could partly explain the different clinical features between clinical phases of infection, the mechanism of which deserves further study.
Conclusion
This study aimed at assessing the distribution of CpG Islands in the genome of Alphapapillomaviruses shows that, in this group of papillomaviruses, CpG Islands are absent in viruses with a definite oncogenic potential. This distribution of CpG Islands in viruses could also serve as a very important element for the identification of oncogenic HPVs. The lack of CpG Islands appears to represent an evolutionary hallmark of HPV related molecular processes. Indeed, the more an HPV is capable of damaging host tissues (oncogenic), the fewer CpG Islands it has, and an observation which could be an evolutionary hallmark for Alphapillomaviruses, characterized by the complete absence of CpG Islands. The findings of this study might provide a foundation for further studies on the role of CpG in determining HPV oncogenic potential.
Acknowledgments
The authors are strongly grateful to all women who accepted to participate in the study. They also express their gratitude to all administrative and traditional officials who facilitate the implementation of this study in each study site.
Funding
“This research received no funding”.
Data Availability Statement
Data are not available upon request due to restrictions (e.g., privacy or ethical). The data presented in this study are available upon request from the corresponding author. Our institution is working on a repository of data.
Author Contributions
Conceptualization, E.E.E.L (Embolo Enyegue Elisee Libert). And GC (Godwe celestion); methodology, E.E.E.L and A.H. (Awalou Halidou); validation, N.D.G (Ndoh Deh Gilbert); formal analysis, E.E.E.L. and B.E.M (Bell Eric Michel); investigation. E.E.E.L; resources, E.E.E.L, A.H, N.D.G; writing-original draft preparation, E.E.EL, N.D.G.; supervision, E.E.E.L; project administration, E.E.EL, B.E.M. funding acquisition, Ourselves. All authors have read and agreed to the published version of the manuscript.
Ethics Approval and Consent to Participate
N/A.
Patient Consent for Publication
N/A.
Competing Interests
The authors declare no conflict of interest.
References
- Chen L, Shi Y, Yang W, Zhang Y, Xie Q, et al. (2018) Differences in Cpg Island distribution between subgenotypes of the hepatitis B virus genotype. Med Sci Monit 24:6781-6794.
[Crossref] [Google Scholar] [PubMed]
- de Sanjose S, Brotons M, Pavon MA (2018) The natural history of Human papillomavirus infection. Best Pract Res Clin Obstet Gynaecol 47:2-13.
[Crossref] [Google Scholar] [PubMed]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H (2004) Classification of papillomaviruses. Virology 324:17-27.
[Crossref] [Google Scholar] [PubMed]
- Egawa N, Egawa K, Griffin H, Doorbar J (2015) Human Papillomaviruses epithelial tropisms, and the development of neoplasia. Viruses 7:3863-3890.
[Crossref] [Google Scholar] [PubMed]
- Galvan SC, Martinez-Salazar M, Galvan VM, Mendez R, Diaz-Contreras GT, et al. (2011) Analysis of CpG methylation sites and CGI among human Papillomavirus DNA genomes. BMC Genomics12:580.
[Crossref] [Google Scholar] [PubMed]
- Hejnar J, Hajkova P, Plachy J, Elleder D, Stepanets V, et al. (2001) CpG island protects Rous sarcoma virus derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc Natl Acad Sci USA 98:565-569.
[Crossref] [Google Scholar] [PubMed]
- Jones SE, Hibbitts S, Hurt CN, Bryant D, Fiander AN, et al. (2017) Human papillomavirus DNA methylation predicts response to treatment using Cidofovir and Imiquimod in vulval intraepithelial neoplasia 3. Clin Cancer Res 23:5460-5468.
[Crossref] [Google Scholar] [PubMed]
- Kirii Y, Matsukura T (1998) Nucleotide sequence and phylogenetic classification of Human papillomavirus type 67. Virus Genes 17:117-121.
[Crossref] [Google Scholar] [PubMed]
- Leitner T, Kumar S (2020) Where did SARS-CoV-2 come from?. Mol Biol Evol 37:2463-2464.
[Crossref] [Google Scholar] [PubMed]
- Ma L, Cong X, Shi M, Wang XH, Liu HY, et al. (2017) Distribution of Human Papillomavirus genotypes in cervical lesions. Exp Ther Med 13:535-541.
[Crossref] [Google Scholar] [PubMed]
- McBride AA (2022) Human papillomaviruses: Diversity, infection and host interactions. Nat Rev Microbiol 20(2):95-108.
[Crossref] [Google Scholar] [PubMed]
- Nishimura A, Ono T, Ishimoto A, Dowhanick JJ, Frizzell MA, et al. (2000) Mechanisms of Human papillomavirus E2 mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J Virol 74(8):3752-3760.
[Crossref] [Google Scholar] [PubMed]
- Niyazi M, Sui S, Zhu K, Wang L, Jiao Z, et al. (2017) Correlation between methylation of Human papillomavirus-16 L1 gene and cervical carcinoma in Uyghur women. Gynecol Obstet 82(1):22-29.
[Crossref] [Google Scholar] [PubMed]
- Upadhyay M, Vivekanandan P (2015) Depletion of CpG dinucleotides in Papillomaviruses and polyomaviruses: A role for divergent evolutionary pressures. PloS One 10:0142368.
[Crossref] [Google Scholar] [PubMed]
- Xia X (2020) Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol Biol Evol 37:2699-2705.
- Xue Y, Wang MJ, Huang SY, Yang ZT, Yu DM, et al. (2016) Characteristics of CpG Islands and their quasispecies of full-length hepatitis B virus genomes from patients at different phases of infection. Springer Plus 5:1630.
[Crossref] [Google Scholar] [PubMed]
- Chen Z, Fu L, Herrero R, Schiffman M, Burk RD (2007) Identification of a novel Human Papillomavirus (HPV97) related to HPV18 and HPV45. Int J Cancer 121:193-198.
[Crossref] [Google Scholar] [PubMed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi