Research Article, J Genet Disor Genet Rep Vol: 11 Issue: 1
Novel Mutation in Slc37a4 Gene Found by Whole-Exome Sequencing in Type I Glycogen Storage Disease (Von Gierke)
Sajad Rafiee Komachali1,2, Zakieh Siahpoosh3 and Mansoor Salehi1,2*
Cellular, Molecular and Genetics Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Medical Genetics Research Center of Genome, Isfahan University of Medical Sciences, Isfahan, Iran
Department of Biology, University of Sistan and Baluchestan, Zahedan, Iran
*Corresponding Author: Mansoor Salehi
Cellular, Molecular and Genetics Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
Tel: +98 3136259017; E-mail: M_salehi@med.mui.ac.ir
Received date: 26 January, 2022; Manuscript No. JGDGR-22-51084;
Editor assigned date: 28 January, 2022; Pre QC No. JGDGR-22-51084(PQ);
Reviewed date: 08 February, 2022; QC No JGDGR-22-51084;
Revised date: 18 February, 2022; Manuscript No. JGDGR-22-51084(R);
Published date: 25 February, 2022; DOI:10.4172/2327-5790.1000204.
Citation: Zakieh Siahpoosh (2022) Novel Mutation in Slc37a4 Gene Found by Whole-Exome Sequencing in Type I Glycogen Storage Disease (Von Gierke) Patient in Iran. J Genet Disor Genet Rep 11:2.
Keywords: Insulin and Cyclic AMP.
Introduction
In recent years the number of disorders that change amino acid synthesis pathway has grown so fast. In addition to the number of disorders that has increase, the associated clinical phenotypes have also expanded. All of these are classified as one of the advances of next generation sequencing diagnostics [1].
Living organisms require a permanent source of energy to support essential processes for life. In our body, most cells get this power from glucose molecule, which enters the cell through glucose transporters and is immediately converted to Glucose-6-Phosphate (G6P) by both hexokinase and glucokinase [2].
Glucose/G6P is metabolized through four pathways mostly, glycolysis, Pentose Phosphate Pathway (PPP), Tricarboxylic Acid (TCA) cycle, and fatty acid synthesis, which produce different substrates that are used in two ways. First way, glycolysis and Oxidative Phosphorylation (OXPHOS) through the TCA cycle provide substrates for Adenosine Triphosphate (ATP) production. Second way, molecules produced at various steps are the substrates utilized as building blocks for macromolecules, such as DNA, RNA, and proteins that are needed for the proliferation, differentiation, and effector functions of cells [2].
Glycogen Storage Disease (GSD) comprises a group of autosomal recessive in born disorders recognized by of the enzyme deficiencies which regulate the synthesis or degradation of glycogen [3,4]. The transformation of glucose into glycogen occurs by chemical reactions carried out by special enzymes, and a deficiency in one of them means accumulation of glycogen, resulting in hereditary disorders known as Glycogen Storage Diseases (GSD) or glycogenosis.
GSDs-I (Von Gierke) are a group of metabolic recognized arising from defects in different components of the G6Pase system [4,5]. Originally in order to the basis of kinetic studies, four different subtypes of GSD-l have been distinguished: GSD-Ia, caused by errors in the catalytic unit; GSD-lb, caused by errors in the G6PT [6]; GSD-Ic, caused by deficient activity of the phosphate/pyrophosphate translocase; and GSD-ld that is proposed to be caused by deficient activity of a putative glucose translocase [7,8].
Molecular genetic evidence has unequivocally demonstrated that mutations in the genes for the G6Pase catalytic unit and the G6PT will result in GSD-la and GSD-lb respectively. Also, many cases of GSD type l-non-a have been diagnosed incorrect [6,7]. In 1994, other subtypes have also been found and labeled as GSD IaSP, Ic, and Id. But with advances in molecular genetic methods, the patients with GSD IaSP carry mutations in G6PC and most patients with GSD Ic and Id carry mutations in the G6PT1 gene SLC37A4. After all, GSD I is now divided into two main subclasses GSD Ia and GSD I non-a, respectively.
The SLC37 family belongs to the largest human Solute-Carrier (SLC) superfamily and includes over 52 gene families, and more than 400 membrane-bound proteins catalyzing the transport of metabolites through biological membranes transmembrane domains [9].
The human SLC37 gene family includes four proteins numbered SLC37A1-4, localized in the Endoplasmic Reticulum (ER) membrane [9,10]. The human G6PT gene (SLC37A4) has been localized to chromosome 11 at 11q23, spans 4.5 kb, contains 9 exons. This gene codes for a highly hydrophobic protein containing 10 trans membrane domains [5,11]. G6PT is expressed in many tissues and in order its tissue specific splicing, there are several variants of it. It is highly expressed in the liver, pancreas, kidney and hematopoietic progenitor cells. In the liver and leucocytes, exon 7 is not expressed. SLC37A4 gene expression is regulated, like that of G6PC, by glucose, insulin and cyclic AMP.
Glycogen Storage Disease (GSD) type I is characterized by hepatomegaly, growth retardation, bleeding tendency, hypoglycemia, hyper lactacidemia, absence of ketosis, hyperuricemia, hyperlipidemia and impaired platelet function. The enzyme defect was first described by Cori and Cori in 1952 [5,12]. Patients with GSD Ia and Ib include and present a nearly similar metabolic phenotype, including hypoglycemia, hepatomegaly, hyperuricemia, lactic acidemia and hyperlipidemia, but GSD Ib patients also manifest neutropenia and myeloid dysfunction and these individuals are susceptible to bacterial infections, aphthous stomatitis, and inflammatory bowel disease. After all, neutropenia is not manifested by all GSD Ib patients [4].
It is notable that disease commonly manifests, between the ages of 3 to 4 months by symptoms of hypoglycemia (tremors, seizures, cyanosis, apnea) and Patients have poor tolerance to fasting that generally improved by a special diet [5]. It is good to know that high rates of consanguineous marriages in Iran provide a desirable context to facilitate finding the homozygous pathogenic mutations [3].
Here, for the first time we report a case which contains NM_001164280: exon9: c.1124-2A>G mutation of the SLC37A4 gene that was identifies by whole exome-sequencing and confirmed by Sanger sequencing. Our aim with this study was to propose that the novel pathogenic variant NM_001164280: exon9: c.1124-2A>G can cause type I glycogen storage disease (Von Gierke).
Materials and Methods
Patient
Present study involves a 4 years old child as a result of a consanguineous marriage that is affected by type I glycogen storage disease (Von Gierke) and have been diagnosed with hepatomegaly, growth retardation, hypoglycemia, hyperlactacidemia, hyperuricemia, hyperlipidemia and poor tolerance to fasting. Local ethics committees received informed consent from the subjected family. Informed consent was obtained from all human adult participants and from the parents or legal guardians of minors in genome laboratory of Isfahan. In this study, internal consent has been prepared, adjusted and available in genome laboratory of Isfahan.
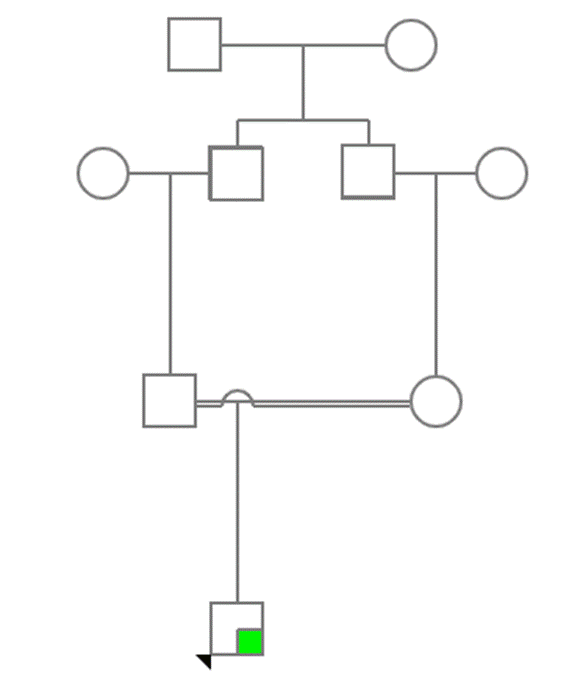
Figure 1: Pedigree of the family.
Mutation analysis
Genomic DNA was extracted from peripheral blood of the patients and their parents, using innuPREP Blood DNA Mini Kit (Analytika Jena,Germany) according to the manufacturer’s protocol.
Samples was under whole-exome sequencing using Genome Analyzer HiSeq 4000 (Illumina, San Diego, CA, USA, 101-bp paired- end reads) following the manufacturer’s instructions. The library was generated using SureSelect XT Library Prep Kit (Agilent Technologies, CA, USA). Sequenced reads were mapped to the GRCH37/UCSC hg19 human reference Genome. To find the disease-causing variation, a filtering pipeline was set up
Exome Sequencing Project (ESP), 1000 Genomes (1000G) and Exome Aggregation Consortium (ExAC) data were used to cross-verify the frequencies of discovered variations.
Sanger sequencing was performed by ABI prism 3730 sequencer (Applied Biosystems, Waltham, MA, USA) to validate the pathogenic mutation and segregation the mutation in this family. Mutation Surveyor program version 4.0.9 was used to analyze the sequences (Soft Genetics, State College, PA).
We employed the 48-well thermocycler device (BioRad) in this reaction. The materials utilized in PCR as well as their concentration and amount were as follows: 6 μL of master (1X), 2 μL of template DNA, 0.5 μL of (10 Pmol) forward primer, 0.5 μL of (10Pmol) reverse primer, and 3.5 L of sterile distilled water (total: 12.5 μL). To prepare the PCR solution, we used 0.2 mL microtubes. We poured the mentioned materials in the tubes and stirred them by pipetting. We amplified the noted gene segment by primers. To amplify the 326-base pair segment, the sequence of forward and reverse primers was ACTCTGAATGCCACTCCACTCTG and TCTCTTCACTCAGCCTTCTTGGAC, respectively.
The utilized primers were manufactured by Tag Copenhagen Co. The 326-base pair segment was amplified in the thermocycler as follows: Cycle 1 for the initial denaturation: once for 5 min at 94°C; cycle 2 including three steps: denaturation, binding the primer to the template strand, and polymerase expansion: 35 times, each for 30 sec at 94, F: 62.19/R: 62.07, and 72°C, respectively; cycle 3 for the final expansion: Once for 10 min at 72°C; cycle 4 for maintaining the products: once at 4°C.
Results
Performing WES on proband identified a novel homozygous NM_001164280: Exon9: c.1124-2A>G mutation of the SLC37A4 gene locates on 11q23 and 118895788 position. Sanger sequencing confirmed heterozygosity of NM_001164280: Exon9: c.1124-2A>G mutation in the proband, suggesting it as the putative disease-causing mutation, and autosomal recessive inheritance pattern type Ib and Ic glycogen storage disease (Von Gierke) (Figure 2).
Our proband was homozygous for glycogen storage disease type Ib and Ic and according to genetic tests performed on the proband’s parents, we found that both of them are heterozygous in this position so autosomal recessive inheritance pattern in this family is confirmed. Through their second pregnancy, this couple returned to genome laboratory of Isfahan for prenatal test. Chorionic Villus Sampling (CVS) was done and the fetus was heterozygous. Contamination of a fetal or cord blood specimen by maternal cells is a potential source of error in diagnostic prenatal testing. In order to the same result of the fetus and its mother; to avoid errors in result, Maternal Cell Contamination (MCC) analysis by using TPO,THO,D3 markers was done and it was confirmed that there were no errors in result so the fetus was truly heterozygous. Although, in order to probabilities of chromosomal abnormalities that increase with maternal age, karyotype test was done on CVS by physician’s request and the result of karyotype was normal.
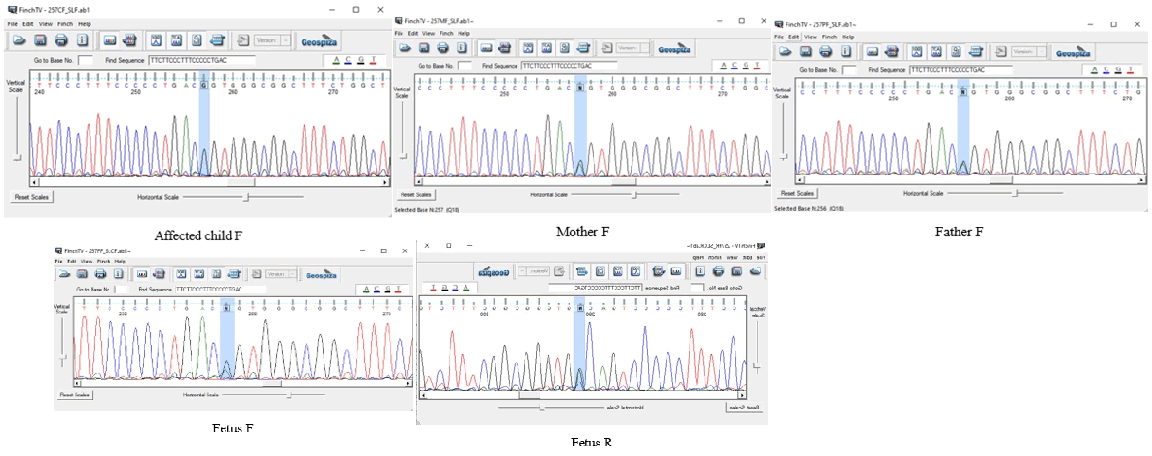
Figure 2: Confirmation sequences of the proband, the fetus and parents.
There is no report of the NM_001164280: Exon9: c.1124-2A>G mutation in SLC37A4 gene in ExAC, 1000G and other control datasets. This variant does not have a gnomAD exomes entry, but its locus is covered in gnomAD exomes as follows. NM_001164280: Exon9: c.1124-2A>G DANN score [13] was 0.9946 and was found as pathogenic mutation in EIGEN predictor [14]. This variant was found as a damaging variant in FATHMM-MKL [15], BayesDel noAF and BayesDel add AF [16] meta-predictors through searching dbNSFP v4 [17]. Also it is notable that NM_001164280: Exon9: c.1124-2A>G mutation was found as a disease causing mutation in mutation taster predictor [18]. In PHRED database, NM_001164280: Exon9: c.1124-2A>G raw score was reported 5.598882 and its PHRED score was reported 34 as we know in 118895788 position [19].
Discussion
Based on the results, our proband that is homozygous for NM_001164280: Exon9: c.1124-2A>G variant as a novel mutation of the SLC37A4 gene is a pathogenic variant that has a great risk factor for type I glycogen storage disease (Von Gierke).
The Glycogen storage disease inherits through autosomal recessive pattern and has an incidence of 1/100,000 to 1/400,000 live births, with up to 80% of the cases represented by subtype Ia [4,5].
In glycogen storage disease patients, clinical therapies consist of dietary treatment, gene therapy and transplantations [9]. Dietary treatment includes preventing hypoglycemia in order to avoid neurological involvement and long-term complications (hepatic, renal, etc.) and also aims to normal growth. Daily caloric intake must be monitored through dietary treatment: Insufficient intake does not correct the metabolic disorder (hypoglycemia, hyperlactacidemia and hyperuricemia) and leads to retarded growth, whereas extra intake increases the glycogen overload, hepatomegaly and hyperlipidemia, and also causes obesity as a result. The diet must provide 60%-65% of total caloric intake from carbohydrates, 10%-15% from proteins and the reminder from fat. Therapeutic adjuvants include vitamin supplements (vitamins D and B1, etc.), calcium (considering limited milk intake), iron in order of presence anemia (after excluding other causes) and allopurinol for hyperuricemia. Hyperlipemia responds partially to dietary treatment, but triglyceride lowering drugs are not needed if the level is below 10 mmol/L. It is notable that Cholesterol lowering drugs are not needed to be taken in young patients because of the low risk of atherogenicity [5].
Gene therapy makes a promising approach to correct all metabolic abnormalities in all GSD patients, although currently it seems not to rectify for myeloid and renal dysfunctions in GSD-Ib patients. Several gene transfer vectors have been developed and tested, both on GSD-Ia and on GSD-Ib animal models including recombinant Adeno Associated Virus (rAAV) vectors [9]. Adeno-Associated Virus (AAV) mediated therapy delivers the transgene to the liver of GSDIa affected mice, and as a well result, achieved long term correction. Only a few experiments were performed in the GSD-Ib models, but an improvement of metabolic defects and myeloid function was reported [5,20].
Conclusion
There are also fine results about transplantations therapies, a recent review showed that 80 GSD-I patients (58 GSD-Ia and 22 GSD-Ib) that have received only liver (52 GSD-Ia and 22 GSD-Ib patients) or combined liver-kidney (6 GSD-Ia patients) transplants have gained normal fasting tolerance and shown better metabolic controls. The six GSD-Ia patients receiving combined liver kidney transplants also lacked further renal disease. However, the effectiveness of just liver transplantation for renal disease in GSD-Ia and GSD-Ib, or myeloid dysfunction in GSD-Ib, remains ambiguous.
To our knowledge this is the first report of NM_001164280: Exon9: c.1124-2A>G mutation discovery and consequently, the results of the present study may be of importance in genetic counseling.
Acknowledgments
The authors would like to thank the participants as well as genome laboratory of Isfahan.
Ethical Considerations
Informed consent was obtained from all human adult participants and from the parents or legal guardians of minors in genome laboratory of Isfahan.
References
- De Koning TJ (2017) Amino acid synthesis deficiencies. J Inherit Metab Dis 40: 609-620.
[Crossref], [Google Scholar], [Indexed]
- Sim SW, Weinstein DA, Lee YM, Jun HSJFl (2020) Glycogen storage disease type Ib: Role of glucose phosphate transporter in cell metabolism and function. FEBS Lett 594: 3-18.
[Crossref], [Google Scholar], [Indexed]
- Eghbali M, Abiri M, Talebi S, Noroozi Z, Shakiba M, et al. (2020) Genotype-phenotype correlation and description of two novel mutations in Iranian patients with Glycogen Storage Disease 1b (GSD1b). Orphanet J Rare Dis 15: 1-11.
[Crossref], [Google Scholar]
- Carlin MP, Scherrer DZ, Tommaso AMAD, Bertuzzo CS, Steiner CE (2013) Determining mutations in G6PC and SLC37A4 genes in a sample of Brazilian patients with glycogen storage disease types Ia and Ib. Genet Mol Biol 36: 502-506.
[Crossref], [Google Scholar], [Indexed]
- Froissart R, Piraud M, Boudjemline AM, Saban CV, Petit F, et al. (2011) Glucose-6-phosphatase deficiency. Orphanet J Rare Dis 6: 1-12.
[Crossref], [Google Scholar], [Indexed]
- Chou JY, Jun HS, Mansfield BC (2013) The SLC37 family of phosphate-linked sugar phosphate antiporters. Mol Aspects Med 34: 601-611.
[Cross Ref], [Google Scholar], [Indexed]
- Foster JD, Nordlie RC (2002) The biochemistry and molecular biology of the glucose-6-phosphatase system1. Exp Biol Med 227: 601-608.
[Cross Ref], [Google Scholar], [Indexed]
- Cunha MV, Gerin I, Schaftingen EV (2000) How many forms of glycogen storage disease type I? Eur J Pediatr 159: 314-318.
[Crossref], [Google Scholar], [Indexed]
- Cappello AR, Curcio R, Lappano R, Maggiolini M, Dolce V (2018) The physiopathological role of the exchangers belonging to the SLC37 family. Front Chem 6: 122.
[Crossref], [Google Scholar], [Indexed]
- Chou JY, Mansfield BC (2014) The SLC37 family of sugar-phosphate/phosphate exchangers. Curr Top Membr 73: 357-382.
[Crossref], [Google Scholar], [Indexed]
- Han SH, Ki CS, Lee JE, Hong YJ, Son BK, et al. (2005) A novel mutation (A148V) in the glucose 6-phosphate translocase (SLC37A4) gene in a Korean patient with glycogen storage disease type 1b. J Korean Med Sci 20: 499-501.
[Crossref], [Google Scholar], [Indexed]
- https://rarediseases.info.nih.gov/diseases/2515/glycogen-storage-disease-type-1b.
- Quang D, Chen Y, Xie X (2015) DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31: 761-763.
[Crossref], [Google Scholar], [Indexed]
- Laza II, McCallum K, Xu B, Buxbaum JD (2016) A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet 48: 214-220.,
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, et al. (2015) An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 31: 1536-1543.
[Crossref], [Google Scholar], [Indexed]
- Tian Y, Pesaran T, Chamberlin A, Fenwick RB, Li S, et al. (2019) Revel and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci Rep 9: 1-6. [Crossref],
- Liu X, Li C, Mou C, Dong Y, Tu Y (2020) dbNSFP v4: Acomprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med 12: 1-8.
[Cross Ref], [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) Mutation taster 2: Mutation prediction for the deep-sequencing age. Nat Methods 11: 361-362.
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces usingPhred. I. Accuracy assessment. Genome Res 8: 175-185.
- Chou JY, Jun HS, Mansfield BC (2015) Type I glycogen storage diseases: Disorders of the glucose-6-phosphatase/glucose-6-phosphate transporter complexes. J Inherit Metab Dis 38: 511-519.
[Crossref], [Google Scholar], [Indexed]
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi 



