Case Report, J Genet Disor Genet Rep Vol: 11 Issue: 6
Novel Variant Documentation of Dystrophic Epidermolysis Bullosa by Whole Exome Sequencing
Priyanka Vishwakarma1*, Mayank Nilay2, Ashish Dubey1, Shashank Upadhyay3, Amit Joshi3, Deepika Kalo1 and Vishal Kumar Mishra4
1Department of Clinical Genomics, Redcliffe Lifetech Private Limited, H-55, 3RD Floor, Sector 63, Gautam Buddha Nagar, Noida, Uttar Pradesh 201301
2Department of Medical Genetics, Post Graduate Institute of Child Health, Sector 30, Noida, Gautam Buddha Nagar, Uttar Pradesh 201303
3Department of Biotechnology, Invertis University, Delhi Lucknow Highway NH-24, Bareilly, Uttar Pradesh 243123
4Department of Bioinformatics, Redcliffe Lifetech Private Limited, H-55, 3RD Floor, Sector 63, Gautam Buddha Nagar, Noida, Uttar Pradesh 201301
*Corresponding Author: Priyanka Vishwakarma, Department of Clinical Genomics, Redcliffe Lifetech Private Limited, H-55, 3RD Floor, Sector 63, Gautam Buddha Nagar, Noida, Uttar Pradesh 201301, India; E-mail: dr.priyanka@redcliffelabs.com
Received date: 04 March, 2022, Manuscript No. JGDGR-22-59140;
Editor assigned date: 07 March, 2022, PreQC No. JGDGR-22-59140 (PQ);
Reviewed date: 21 March, 2022, QC No. JGDGR-22-59140;
Revised date: 03 May, 2022, Manuscript No. JGDGR-22-59140 (R);
Published date: 10 May, 2022, DOI:10.4172/2327-5790.1000015
Citation: Vishwakarma P, Nilay M, Dubey A, Upadhyay S, Joshi A, et al. (2022) Novel Variant Documentation of Dystrophic Epidermolysis Bullosa by Whole Exome Sequencing. J Genet Disor Genet Rep 11:6.
Abstract
Epidermolysis Bullosa (EB) is an inherited disorder. It involves the heterogeneous group of the rare genetic dermatoses which are characterized by the mucocutaneous brittleness and the blister development and it is often inducible by the minimal trauma. A wide-ranging phenotypic diversity has been defined, with possibly severe extra-cutaneous appearances, morbidity and the mortality in some cases. In this study we have documented a case of the Epidermolysis bullosa with a novel variant in COL7A1 detected through whole exome sequencing. This novel mutation has been authenticated by the Sanger sequencing. This case highlights the importance of whole exome sequencing for confirmatory molecular diagnosis and adds a novel variant to the genotypic spectrum of COL7A1 from the Indian Subcontinent.
Keywords: Skin lesions; Epidermolysis; whole exome sequencing; Sanger sequencing; Novel Variant
Abbreviations
HPO: Human Phenotype Oncology; dbSNP: Database Single Nucleotide Polymorphism; gnomAD: The Genome Aggregation Database; LRT: Likelihood Ratio Test; SIFT: Sorting Intolerant from Tolerant; OMIM: Online Mendelian Inheritance in Man; HGMD: Human Gene Mutation Database; AFs: Anchoring Fibrils
Introduction
Epidermolysis Bullosa (EB) is a collection of genetic skin illnesses that originate in the skin as blisters with skin erosion on minimal trauma. Aberrations of the macromolecules, which anchor the dermis to the epidermis leads to the reduced cohesion of the skin layers, blister formation, and fragility. The ruthlessness of the skin appearances can be highly variable and is dependent on the mode of inheritance and the underlying mutation. In the people with EB, blisters form in retort to minor injuries or friction, such as rubbing or scratching. There are four main types of the EB, which are categorized based on the complexity, or level of blister formation, EB simplex, dystrophic EB, functional EB and kindler syndrome [1]. Most of the patients presenting to a genetic/dermatological unit belongs to the DEB subtypes owing to the augmented morbidity accompanying with this subtype [2].
EB may then be further categorized on the basis of its severity and by the help of specific symptoms, such as the distribution and whether the parts of the body other than the skin are exaggerated in the affected individual. The specific sub-types may then be determined on the basis of categorizing the exact protein that is defective in a person with the EB. Mutations in the 14 genes have been identified till date as the causes of the EB in the human beings [3,4]. The identifications of EB may be done by the tests performed on a skin biopsy, or when by the possible, genetic testing. Even the direct genetic testing is feasible in pediatric populations owing to avoidance of the painful skin biopsies and the advantage of the molecular confirmation. A person with EB may be mildly or severely affected, and the disease can range from being a minor inconvenience requiring modifying activities, to completely disabling and even fatal in the some cases.
EB may be caused by variations (the changes called mutations) in at least 18 genes that play vital roles in the structure, integrity, and the repair of the skin. Inheritance pattern may be autosomal dominant or autosomal recessive depending on the type and the subtype of the EB a person has managing EB involves a multidisciplinary team of the health care workers comprising a dermatologist, EB nurse who specializes in the care of wound, professional therapist, nutritionist, Geneticist and the social worker [5]. Management should be individualized for each person depending on their age, severity of the symptoms, and the associated complications[6].
Presently there is no any specific curative therapy for most forms of the EB; the extensive clinical research regarding the potential treatments is ongoing. At this time, management is frequently supportive. Monitoring for the complications with the laboratory testing and the imaging studies is also important, although the frequency of these tests will vary depending on the type of EB and severity in each person. New-borns with EB should be taken care of in a neonatal or pediatric unit that has the expertise, staffing, and resources necessary to manage severe skin erosions and potential complications.
In this paper we used the utility of Whole Exome Sequencing (WES) which is a widely used Next Generation Sequencing (NGS) method that involves sequencing the protein-coding regions of the genome [7]. Owing to its high throughput nature and the ability to instantaneously sequence the multiple genes, NGS, specifically the Whole Exome Sequencing (WES), has modernized the molecular approach and it’s provided a fast and well-versed diagnostic strategy in a number of genodermatoses [8,9]. WES is a proficient approach to selectively sequence the coding regions (exons) of the genome to discover the rare or common variants associated with a disorder or phenotype [10,11].
Case Presentation
The subject is a 3 years old, female (non-consanguineous family), who was brought to the clinician with complaints of failure to thrive, skin blisters and peeling of skin on the trunk and limbs with minimal friction since birth. She also had milia and pseudo syndactyly of fingers and toes. Oral mucosal lesions and nail dystrophy were evident on examination. There was no similar complaint in the family. A provisional diagnosis of the Dystrophic Epidermolysis bullosa was made and the Whole exome sequencing was performed to ascertain the exact molecular diagnosis as the parents did not give consent for skin biopsy.
Test methodology
The DNA extraction from the blood was used to perform the targeted gene capture by the help of a custom capture kit. The libraries were sequenced to mean >100X coverage on Ilumina sequencing platform. The sequences obtained are aligned to human reference genome (GRCh37/hg19) and variant analysis was performed using set of Bioinformatics Pipeline.
Variant exploration
Golden Helix VarSeq 2.2.0 is a clinical genomics interpretation and reporting platform from Golden Helix. The variant annotation engine includes algorithms to identify variant impact on gene using both public content (ClinVar, HPO, links to dbSNP, gnomAD and in-silico predictors-GERP++, PhyloP, PhyloP LRT, SIFT and PolyPhen2. VarSeq allows quick filtering and evaluation of variants. The clinically relevant variants were annotated using published variants in literature and a set of diseases databases-dbSNP, ClinVar, OMIM and HGMD. Common variants are filtered based on allele frequency in gnomAD. Only nonsynonymous and splice site variants found in the exome panel consisting of specific set of genes were used for clinical interpretation. Silent variations that do not result in any change in amino acid in the coding region are not reported.
Results
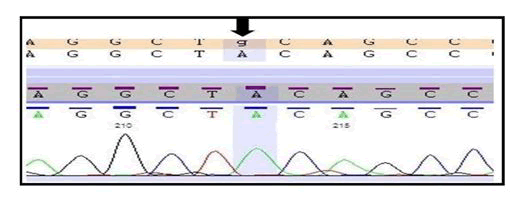
In this study, we found a novel variant (c.1966C>T; p. Gln656Ter) in COL7A1 by the help of widely used technique whole exome sequencing and also this variant was successfully validated by the gold standard method Sanger sequencing (Figure 1).
The individual carries two copies of a nonsense variant in the COL7A1 gene which is predicted to cause the premature truncation of the protein. This variant seems to be novel as it has not been previously reported in the scientific literature and in the databases. Since, this variant is prophesied to produce a truncated protein which might result in the loss of the function and other truncating variants in this gene are also known to cause similar phenotype, therefore, this variant has been labelled as pathogenic (disease causing) (Table 1).
| Gene | Chromosomal Coordinates | Exon | Variant | Zygosity | Disease | Significance (ACMG Classification) | Inheritance | Coverage |
|---|---|---|---|---|---|---|---|---|
| COL7A1 (NM_000094.3) | chr3:48627730: G>A | 15 | c.1966C>Tp. Gln656Ter | Homozygous | Epidermolysis bullosa dystrophica | Pathogenic (PVS1,PM2,PP3) | AutosomalRecessive | 111X |
*The nucleotide sequence numbering for COL7A1 is as per NM_000094.3
Table 1: A novel variant identified by WES (Whole Exome sequencing) in COL7A1 gene.
According to the ACMG (American College of Medical Genetics) guidelines [12]. This variant is also present in the important Fibronectin III domain of COL7A1 protein.
Discussion
DEB is an extremely rare subtype of EB which is caused by mutation of the COL7A1 gene. EB is characterized by blistering with scarring predominantly which can occur in childhood or the adulthood stage [13]. Initially, the diagnosis of DEB was suspected in patient because it overlapped with other subtypes with respect to symptoms. Biopsy is very important to confirm EB [14,15]. But we could not obtain biopsy from the patient because they did not provide the consent. In worldwide study there are lots of studies reported in the different exons of COL7A1 gene related with EB and other similar disease but in our study we are reporting a case of EB with novel mutation in the exon 15 of COL7A1 gene [16-20].
Gene COL7A1 present on the locus of chromosome 3p21.1 position and it spans approximately 32 kb and encompasses 120 exons. COL7A1 gene encrypts a type-VII collagen which is a major structural component of AFs (Anchoring Fibrils). It is responsible for the cohesion of the epidermis and dermis. Kon et al. reported the first mutation in the COL7A1. To date, more than 730 pathogenic mutations have been detected within the COL7A1 gene in different variants of the DEB. Almost all of them are family-specific, although a small number of recurrent mutations have been reported. However, different case lead to the same subtypes of DEB [16-18].
The Dystrophic EB is a severe skin disorder commonly present since birth and categorized by recurrent blistering at the level of the sub-lamina densa beneath the cutaneous basement tissue membrane. The Recessive DEB has severe phenotype with generalized involvement; scarring and patients may have contractures of the hands, feet, and the joints. Patients may also develop strictures of the gastrointestinal tract from the mucosal involvement, which leads to dysphagia-poor nutrition-failure to thrive. The affected individuals also have an augmented risk of the developing aggressive squamous cell carcinoma (OMIM 226600). The onset of these clinical features has been evident in the early childhood, but in some cases it is delayed till the second or third decade of the life.
Despite well-characterized genetic studies from different ethnic backgrounds, identifying several recurrent and region-specific mutations, molecular diagnosis of DEB is still challenging [19]. Preceding to the initiation of Next Generation DNA Sequencing (NGS) technologies, molecular diagnosis of DEB was based on traditional Sanger sequencing of either the hot-spot regions or the entire COL7A1 gene, requiring more than 70 primer sets; a process that is tedious, time consuming and expensive. Due to its high throughput nature and ability to simultaneously sequence multiple genes, NGS, especially Whole Exome Sequencing (WES), has revolutionized the molecular approach and provided a fast and efficient diagnostic strategy in several genodermatoses. In view of the scarcity of, and need for, molecular studies for DEB in India, the aim of this study was to explore the probable of WES in the correct diagnosis of the patient with DEB, as part of a larger effort in understanding the mutational spectrum of patient with EB in Indian study. This study by using the utility of WES diagnosis is clearly promising and will serve as a basis for the future large-scale molecular studies of EB patients in the Indian population.
Conclusion
This case report highlights the advantage of the Whole Exome Sequencing in the exact molecular diagnosis with the high diagnostic yield in Genodermatoses. The meticulous molecular diagnosis helps in the management, prognosis, and prenatal diagnosis and also as a potential source for the therapeutic trials in the area of precision medicine and therapy.
Authors contribution
PV is working as Scientist, have made substantial contributions to conception and design, which worked and compiled the data and drafted this manuscript. MN is a Medical Geneticist, have been involved in drafting the manuscript or revising it critically for important intellectual content. SD is a Scientist who reviewed the study. AD did a substantial contribution in finalizing and critical review of the manuscript. DK is working as a Scientist who gave her valuable input or acquisition of data, or analysis and interpretation of data during the manuscript preparation. AJ is a scientist who reviewed the study thoroughly. VKM is bioinformatics Scientist, who provided his major input during data analysis.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Laimer M, Prodinger C, Bauer JW (2015)Hereditary epidermolysis bullosa. J Dtsch Dermatol Ges 13:1125-1133.
- Nilay M, Saxena D, Mandal K, Moirangthem A, Phadke SR (2021) Novel pathogenic variants in an Indian cohort with epidermolysis bullosa: Expanding the genotypic spectrum. Eur J Med Genet 64:104345.
- Fine JD, Eady RA, Bauer EA, Bauer JW, Bruckner-Tuderman L, et al. (2008) The classification of inherited epidermolysis bullosa (EB): Report of the Third International Consensus Meeting on Diagnosis and Classification of EB. J Am Acad Dermatol 58:931-950.
- Fine JD (2010) Inherited epidermolysis bullosa: recent basic and clinical advances. Curr Opin Pediatr 22:453-458.
- Murrell DF, Intong-Wheeler L (2018) Overview of the management of epidermolysis bullosa. UpToDate Waltham.
- Rani S, Gupta A, Bhardwaj M (2019) Epidermolysis bullosa pruriginosa: A rare entity which responded well to thalidomide. Dermatol Ther 32:e13035.
- Takeichi T, Liu L, Fong K, Ozoemena L, McMillan JR, et al. (2015) Whole-exome sequencing improves mutation detection in a diagnostic epidermolysis bullosa laboratory. Br J Dermatol 172:94-100.
- Tenedini E, Artuso L, Bernardis I,Artusi V, Percesepe A, et al. (2015) Amplicon-based next-generation sequencing: an effective approach for the molecular diagnosis of epidermolysis bullosa. Br J Dermatol. 173:731-738.
- Biesecker LG (2010) Exome sequencing makes medical genomics a reality. Nat Genet 42:13-14.
- Richards S, Aziz N, Bale S, Bick D, Das S, et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405-424.
- Rizzo C, Anandasabapathy N, Walters RF, Rosenman K, Kamino H, et al. (2008) Pretibial epidermolysis bullosa. Dermatol Online J 14:26.
- Kon A, Nomura K, Pulkkinen L, Sawamura D, Hashimoto I, et al. (1997) Novel glycine substitution mutations in COL7A1 reveal that the Pasini and Cockayne-Touraine variants of dominant dystrophic epidermolysis bullosa are allelic. J Invest Dermatol 109:684-687.
- Kon A, McGrath JA, Pulkkinen L, Nomura K, Nakamura T, et al. (1997) Glycine substitution mutations in the type VII collagen gene (COL7A1) in dystrophic epidermolysis bullosa: implications for genetic counseling. J Invest Dermatol 108:224-228.
- Christiano AM, Fine JD, Uitto J (1997) Genetic basis of dominantly inherited transient bullous dermolysis of the newborn: a splice site mutation in the type VII collagen gene. J Invest Dermatol 109:811-814.
- Christiano AM, Hoffman GG, Zhang X, Xu Y, Tamai Y, et al. (1997) Strategy for identification of sequence variants in COL7A1 and a novel 2-bp deletion mutation in recessive dystrophic epidermolysis bullosa. Hum Mutat 10:408-414.
- Varki R, Sadowski S, Uitto J, Pfendner E (2007) Epidermolysis bullosa. II. Type VII collagen mutations and phenotype-genotype correlations in the dystrophic subtypes. J Med Genet 44:181-192.
- Yenamandra VK, Vellarikkal SK, Chowdhury MR, Jayarajan R, Verma A, et al. (2018) Genotype-Phenotype Correlations of Dystrophic Epidermolysis Bullosa in India: Experience from a Tertiary Care Centre. Acta Derm Venereol 98:873-879.
- Mellerio JE, Salas-Alanis JC, Talamantes ML, Horn H, Tidman MJ, et al. (1998) A recurrent glycine substitution mutation, G2043R, in the type VII collagen gene (COL7A1) in dominant dystrophic epidermolysis bullosa. Br J Dermatol 139:730-737.
- Chuang GS, Martinez-Mir A, Yu HS, Sung FY, Chuang RY, et al. (2004) A novel missense mutation in the COL7A1 gene underlies epidermolysis bullosa pruriginosa. Clin Exp Dermatol 29:304-307.
- Lin Y, Chen XJ, Liu W, Gong B, Xie J, et al. (2012) Two novel mutations on exon 8 and intron 65 of COL7A1 gene in two Chinese brothers result in recessive dystrophic epidermolysis bullosa. PLoS One 7:e50579.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi