Research Article, J Pharm Sci Emerg Drugs Vol: 7 Issue: 2
Mutational Mosaicism in Breast Cancer
1Center for Medical Innovation, Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea
2Seoul National University Bio-MAX/N-Bio, Seoul, South Korea
*Corresponding Author : Ryong Nam Kim
Seoul National University Bio-MAX/NBio, Seoul, South Korea
E-mail: ryongnamkim@gmail.com
Received Date: August 28, 2019; Accepted Date: September 13, 2019; Published Date: September 25, 2019
Citation: Kim RN (2019) Mutational Mosaicism in Breast Cancer. J Pharm Sci Emerg Drugs 7:2.
Abstract
In recent years, a huge number of mutational variants had been identified in breast cancer by next-generation sequencing technologies. Even though a considerable portion of them are variants with low variant allele fraction (<30%), which could give rise to suspicion among us regarding whether they might be false or true, recent pioneering studies have begun to corroborate that a certain amount of them are true variants associated with mutational mosaicism. In this study, for the first time, we present pathogenic mutational mosaicism in breast cancer by carrying out comprehensive analysis of large-scale somatic mutation databases far beyond a limited scale of individual cohorts. We identified 23 pathogenic and likely pathogenic mutations with low variant allele fraction (≤ 30%). Of them, there are 8 TP53, 3 PIK3CA, 2 KRAS and 2 GNAS mutations, and one each of SLC25A19, OTC, PACS1, FLG, NCF4, UROS, MLC1, and LTBP2 mutations. For 9 of the mosaic mutations, their variant allele fractions are more than 50% in clinical breast cancer samples compared with those in the normal blood samples, suggesting their contribution to predisposition for carcinogenesis. Three TP53 mosaic mutations (pY220S, p.R273C and p.V272M), and UROS p.L4F could affect directly or indirectly post-translational modifications (phosphorylation, methylation and acetylation). In addition, our protein structural analysis revealed that 4 pathogenic mosaic mutations (p.S241C, p.R273C and p.R248W in p53, and PIK3CA p.E545K) could reside on contact surfaces for protein-protein interactions, consequently affecting the interactions essential for DNA repair pathway. Recurrence free survival analysis showed that expression level of the genes associated with mosaic mutations could be significantly related with patients’ survival. Furthermore, our analysis of somatic variant databases revealed that the 23 pathogenic mosaic mutations might make pivotal contribution to predisposition for carcinogenesis in not only breast cancer but also diverse other cancer types. Taken together, our result presents pathogenic mosaic mutations associated with breast cancer predisposition, which will help clinicians, clinical oncologists and tumor biologists predict breast cancer predisposition, diagnose breast carcinogenesis, choose therapeutic treatment options and elucidate oncogenic mechanisms in the upcoming years.
Keywords: Breast cancer; Mutational mosaicism; Heterozygosity
Keywords
Breast cancer; Mutational mosaicism; Heterozygosity
Introduction
Next generation sequencing (NGS) has revolutionized the process for the discovery and analysis of cancer-causing mutations in human cancer genomes [1]. In addition, recent rapid reduction in the costs for NGS and NGS-based targeted sequencing has enabled not only the acceleration of such discovery but also the beginning of new era called targeted deep sequencing [2].
The NGS-based deep sequencing era has begun to insinuate that the mosaic mutations, which had been previously regarded as inherited mutations rarely occurring in a pattern biased to particular sites in human body, might be not rare phenomenon, but generalized fact for patients with diseases [3]. Nevertheless, a majority of previously published papers reporting deleterious germline and somatic mutations in breast cancer are still ignoring those harmful mutations with low variant allele fraction, often removing them from finally annotated mutation lists.
The major reason of why those mutations with low variant allele fraction were often ignored was that they could not be easily validated by conventional Sanger sequencing method. However, given that recently developed approaches, including digital differential polymerase chain reaction (ddPCR) and barcoded deep sequencing, have accurately validated those mutations with low variant allele fraction in cancer samples [4], these research and clinical fields are entering new stage at which such ignorance may not be allowed.
Another important reason about why this issue should be raised with a priority in dealing with breast cancer is that among so far known cancer types, breast and ovarian cancers might be caused or predisposed by hereditary or germline mutations to the highest extent [5-8]. In contrast to somatic mutations whose variant allele fraction values might not be accurately assessed due to tumor purity issue, the assessment of the variant allele fractions for the hereditary or germline mutations in normal blood or normal tissue reflects relatively correct values because of no purity issue.
That’s why we should not neglect the mosaic mutations with low variant allele fractions in blood or normal tissue, in particular in case of the pathogenic variants. In this regard, the exome aggregation consortium (ExAC) database may be a good resource for exploring such pathogenic mosaic mutations with low variant allele fraction.
Unlike biallelic somatic mutations, loss of heterozygosity (LOH) event showing the somatic mutation in the allele corresponding to a germline mutation in another allele is very typical to breast and ovarian cancer types [9]. However, it has been recently reported that haploinsufficiency event alone could increase susceptibility to carcinogenesis even in the absence of the LOH event of the deleterious pathogenic mutation [10,11].
Given a fact that the VAF levels of deleterious mosaic mutations could often increase up to a degree more than or around 50% in somatic tumor tissues, those pathogenic mosaic mutations might become pivotal contributors to carcinogenesis and its predisposition. In this paper, we show, for the first time, mutational mosaicism common in breast cancer and several other cancer types by exploring public germline and somatic mutation databases in order to address the issue of mutations with low VAF raised recently by this research and clinical community.
Materials and Methods
In order to obtain the mosaic pathogenic somatic mutation data, we intersected Exome Aggregation Consortium (ExAC) database and non-TCGA ExAC database with COSMIC and BRCA TCGA somatic mutation data and ClinVar database. In order to get insights of whether the mosaic mutations are onto the interface for the proteinprotein interaction, we had used the interactome INSIDER software. To get insights of whether the mosaic mutations could be influential to the post-translational modifications, we had used ActiveDriverDB. To perform cox hazard ratio survival analysis, we had used the preprocessed microarray dataset for breast cancer patients from the previously published paper (Gyorffy et al. 2010).
The high and low expressions are defined as values above and below expression median for a given gene, respectively. R programming language had been used for obtaining the mosaic somatic pathogenic mutation data through intersecting among diverse databases.
Results
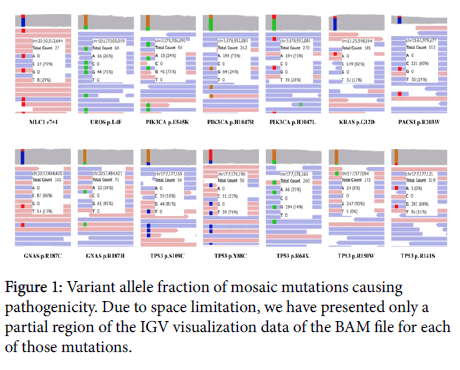
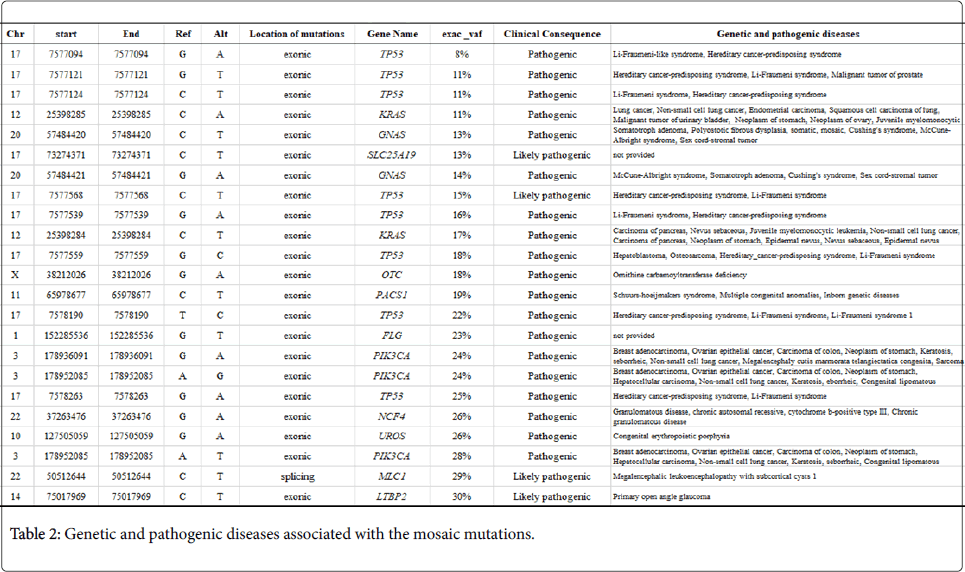
Discovery of breast cancer mosaic mutations through exploring germline and somatic mutation databases. In order to discover breast cancer-causing pathogenic mosaic mutations, we performed a comparison between breast cancer somatic and germline mutation data in the COSMIC database and the Exome Aggregation Consortium database. To guarantee the scientific reliability of our results, we had narrowed down a primarily chosen mosaic mutation list to a finally selected list (23 mosaic mutations with VAF ≤ 30%), in which there are only mosaic mutations being co-occurred in both germline and somatic variant databases and also known as pathogenic or likely pathogenic mutations in Clinvar database (Figure 1 and Table 1). To compare the genomic positions of the variants between the COSMIC database and the Exome Aggregation Consortium database, we have applied R programming.
Even though those variants had been previously known as pathogenic mutations, little is known about whether they might be occurring as mosaic mutations. In order to address this issue, we had analyzed IGV data of BAM files generated by whole exome nextgeneration sequencing of the clinical samples harboring those mutations in the Exome Aggregation Consortium database. As shown in (Figure 1), we have confirmed by their IGV data that those mosaic pathogenic mutations had variant allele fraction ≤ 30%.
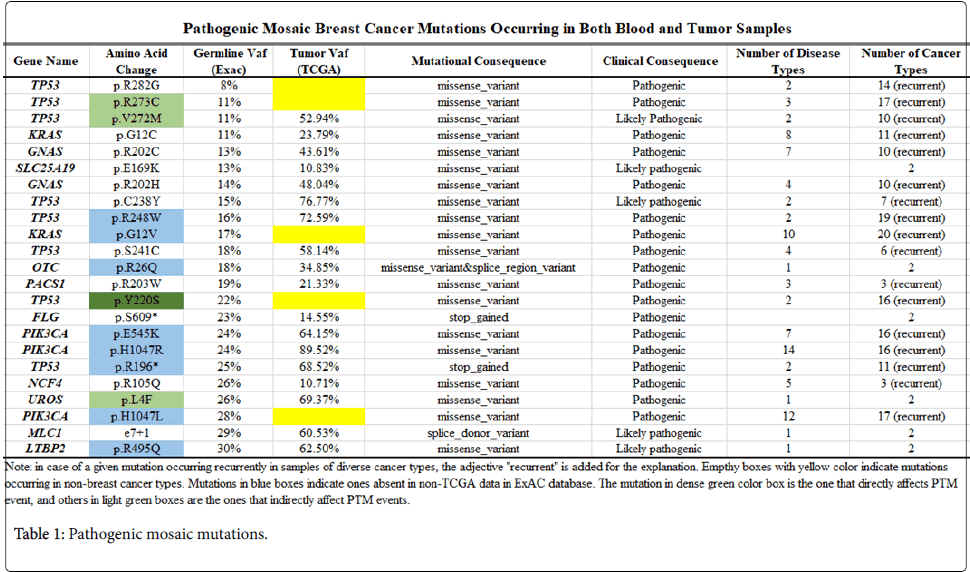
Among the 23 mosaic pathogenic mutations, there are 8 TP53 mutations (p.R282G, p.R273C, p.V272M, p.C238Y, p.R248W, p.S241C, p.Y220S, and p.R196*), 2 PIK3CA mutations (p.E545K, p.H1047R and p.H1047L), 2 KRAS mutations (p.G12C and p.G12V), 2 GNAS mutations (p.R202C and p.R202H), SLC25A19 p.E169K, OTC p.R26Q, PACS1 p.R203W, FLG p.S609*, NCF4 p.R105Q, UROS p.L4F, MLC1 e7+1 (splicing donor mutation in intron), and LTBP2 p.R495Q.
To identify whether those pathogenic mosaic mutations might be closely associated with the susceptibility and causation of breast cancer, we have considered their somatic VAFs in TCGA breast cancer clinical samples (Table 1). Variation nucleotides for the five mosaic mutations (TP53 p.R282G, TP53 p.R273C, KRAS p.G12V, TP53 p.Y220S, and PIK3CA p.H1047L) are replaced with alternative nucleotides at the corresponding genomic positions in TCGA clinical samples, even though the alternative variants also had been known as pathogenic somatic mutations.
The somatic VAFs corresponding to 12 of the remaining 18 pathogenic mosaic mutations increased in the clinical breast cancer samples, compared with the VAFs in their blood samples. In particular, the somatic VAFs corresponding to 10 pathogenic or likely pathogenic mosaic mutations were over 50%, suggesting their likely contribution to breast predisposition to tumorigenesis in clinical samples.
Another important point we should address here is that those twenty-three pathogenic or likely pathogenic mosaic mutations could occur in patients with not only breast cancer but also diverse other cancer types (range: from 2 to 20 cancer types) (Table 1). Furthermore, of the 23 mutations, seventeen occurred recurrently in multiple tumor samples in individual cancer types. This suggests that the cancer causing and predisposition mechanisms by those pathogenic or likely pathogenic mosaic mutations might be shared by diverse cancer types.
Change of PTM sites by mosaic pathogenic mutations
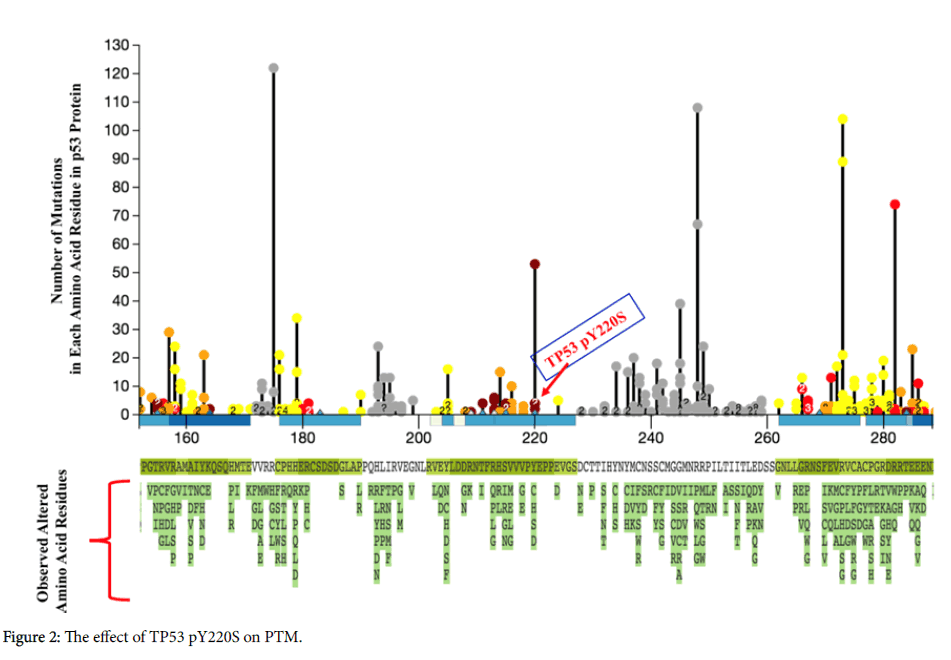
In order to check whether those pathogenic mosaic mutations might change or affect the post-translational modification (PTM) sites that are important in providing proteins with proper functional activities, we compared their genomic positions with PTM sites database. We identified that the pathogenic mosaic mutation TP53 pY220S could inhibit directly the phosphorylation modification of the position Y220 by AURKA, AURKB and AURORA A. Also, this mosaic mutation might affect indirectly the phosphorylation and methylation modifications of the positions S215 and R213 resided very near to Y220, respectively (Figure 2).
In addition, pathogenic mosaic mutations TP53 p.R273C and TP53 p.V272M could affect indirectly in distal manner the phosphorylation of the position S269. Furthermore, we identified that the pathogenic mosaic mutation UROS p. L4F could affect indirectly in distal manner the acetylation of the position K7. Those results suggest that the mosaic pathogenic mutations could be implicated in causing pathogenicity by rewiring crucial pathways through affecting directly or indirectly post-translational modifications including phosphorylation, methylation and acetylation that are essential for downstream cell signaling.
The Figure shows the post-translational modification landscape in the region surrounding the residue Y220 in the p53 protein amino acid sequence. Red, yellow, orange, black and red-brown circles indicate network-rewiring, distal, proximal, no and direct effects on PTM by mutations, respectively. In the horizontal axis showing numbering for amino acid residues in p53, light and dense blue bars indicate protein domain regions undergoing phosphorylation and acetylation or ubiquitination, respectively, and light and dense green bars the domain regions undergoing methylation and ubiquitination, respectively. Region without bar indicates region undergoing none of PTM. Numbers in the circles indicate how many of different mutations with same occurring count overlap at the same residue.
For instance, as shown in the Figure 2, there are three different circles on the residue 220. However, there are four different mutations TP53 pY220C, TP53 pY220H, TP53 pY220S and TP53 pY220D at the residue 220 with counts of 53, 4, 4, and 2 (calculated in TCGA clinical patients), respectively. That is, on the residue Y220, going from the highest to the lowest along the vertical line, the first circle represents TP53 pY220C, and the second circle with the number 2 indicates both TP53 pY220H and TP53 pY220S, and the third one is TP53 pY220D.
Breakdown of DNA repair pathways by the pathogenic mosaic mutations
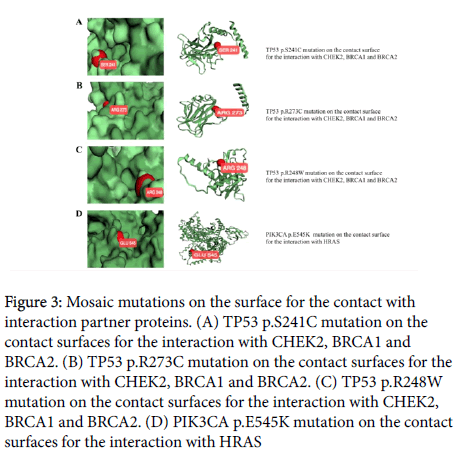
In order to elucidate how the pathogenic mosaic mutations might play pivotal roles in causing predisposition to carcinogenesis, we have performed an analysis about on which surface of the threedimensional protein structure the pathogenic mosaic mutations could reside by using the interactome INSIDER software. As shown in the (Figure 3), we have identified that each of the three mosaic pathogenic mutations TP53 p.S241C, TP53 p.R273C and TP53 p.R248W could reside in unhidden states on the contact surface for physical interactions between p53 and each of CHEK2, BRCA1 and BRCA2 proteins, respectively. It had been known that BRCA2 protein could interact physically and functionally with p53 in order to take part in the DNA repair pathway for maintaining the genomic integrity [12,13] In addition, the fact that BRCA1 protein could play a role as a p53 coactivator had been elucidated by revealing an interaction complex between the two proteins using coimmunoprecipitation technique in a previous investigation [14]. Furthermore, in response to DNA damage, p53 had been reported to undergo C-terminal phosphorylation by CHEK2 [15]. The p53 protein harboring the above-mentioned three pathogenic mosaic mutations might not properly interact with BRAC1, BRCA2 and CHEK2 proteins, subsequently resulting in a breakdown or unfavorable rewiring of DNA repair pathway and consequently causing breast carcinogenesis.
Figure 3: Mosaic mutations on the surface for the contact with interaction partner proteins. (A) TP53 p.S241C mutation on the contact surfaces for the interaction with CHEK2, BRCA1 and BRCA2. (B) TP53 p.R273C mutation on the contact surfaces for the interaction with CHEK2, BRCA1 and BRCA2. (C) TP53 p.R248W mutation on the contact surfaces for the interaction with CHEK2, BRCA1 and BRCA2. (D) PIK3CA p.E545K mutation on the contact surfaces for the interaction with HRAS
We also identified that the pathogenic mosaic mutation PIK3CA p.E545K could reside on the contact surface for the interaction between PIK3CA and HRAS proteins (Figure 3). It had been known that activated RAS protein could stimulate PI3-kinase in addition to Raf in order to induce transformation of mammalian cells and cytoskeletal reorganization [16]. In contrast to the above-mentioned three TP53 mosaic mutations that could be involved in rewiring or weakening the DNA repair pathway, the pathogenic mosaic mutation PIK3CA p.E545K might enhance the transformation effect of the mammalian cells by further reinforcing the interaction on the contact surface between the PIK3CA and HRAS proteins [17].
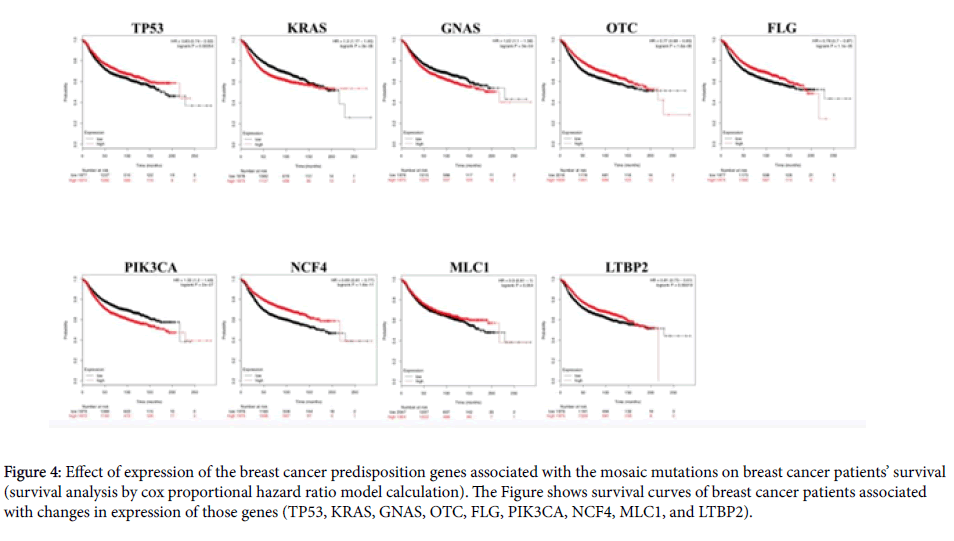
Effect of expression of the breast cancer predisposition genes associated with the mosaic mutations on patient survival
In order to elucidate how the expression level of the abovementioned breast cancer genes in about 1800 clinical breast cancer samples could affect the survival of the patients, we had performed cox survival analysis [18]. For instance, the mutation TP53 p.R196* causing a truncation of p53 protein structure might decrease expression of genuine p53 in cancer tissue of the patients (Table 1). A recent report on the germline and somatic mutations in clinical cancer patients has shown that such truncation mutations in tumor suppressor genes could dramatically decrease expression of those genes in clinical cancer samples [19]. If so, whether such expression changes might affect survival of cancer patients should be elucidated. As shown in (Figure 4), we identified that recurrence-free survival of breast cancer patients with low expression of p53 could decrease significantly, compared with patients with high expression of p53, during more than 200 months (logrank P value=0.00054).
The pathogenic mosaic mutation FLG p.S609* causing a truncation of a major portion of FLG exon 3 harboring Filaggrin as a functional domain, could cause a reduction in the expression level of this gene’s genuine transcript. It had been well known that FLG mutation could be closely associated with the causation of breast cancer [20]. We also identified that the recurrence-free survival of patients with breast tumor showing low expression of FLG could decrease, compared to patients with high expression of FLG, during over 170 months logrank P value=0.000011).
Figure 4: Effect of expression of the breast cancer predisposition genes associated with the mosaic mutations on breast cancer patients’ survival (survival analysis by cox proportional hazard ratio model calculation). The Figure shows survival curves of breast cancer patients associated with changes in expression of those genes (TP53, KRAS, GNAS, OTC, FLG, PIK3CA, NCF4, MLC1, and LTBP2).
In addition, we analyzed relationship between the recurrence-free survival of breast cancer patients and expression levels of genes associated with the other mosaic pathogenic mutations. As shown in (Figure 4) the changes in expression level of KRAS, GNAS, OTC, PIK3CA, NCF4, MLC1 and LTBP2 genes, could affect significantly the survival of breast cancer patients (logrank P values of 2 × 10-6, 3 × 10-4, 1.6 × 10-6, 2 × 10-7, 1.6 × 10-11, 0.054, 0.00019, respectively).
Interestingly, in case of oncogenic genes, such as KRAS, GNAS and PIK3CA, their high expression could be associated with lower survival compared with patients with their low expression. This phenomenon is not discrepant with the recent report about germline and somatic pathogenic mutations in oncogenes in cancer patients, according to which expression of oncogenes with pathogenic variants could increase significantly, promoting carcinogenesis [19]. In contrast, in case of tumor suppressor genes, such as TP53, breast cancer patients with low expression of TP53, could have poor prognosis, compared to those with its high expression. This also is not discrepant with the result in the recent report, according to which expression of tumor suppressor genes with truncation or loss-of-function mutation could decrease significantly in cancer samples.
Discussion
In this study, we have presented pathogenic mosaic mutations, which could concurrently and recurrently occur in both normal blood and breast cancer tissue, as well as other diverse cancer types, including stomach, oesophageal, ovarian, malignant melanoma, liver, lung, pancreatic, colorectal, prostate and glioma cancers (Table 2).
Until now, scientific society investigating clinical cancer had largely ignored mutations with low variant allele fraction less than 30%, considering them as erroneous variant callings. Given the fact that the exome aggregation consortium had used the strictly tight threshold (99.6% sensitivity) to discover real variants and also our chosen pathogenic variants were overlapped with the previously known Clinvar pathogenic or likely pathogenic mutation positions, mosaic mutational statuses of the chosen pathogenic mutations in this study are very reliable.
Recent hot debate issue regarding viewpoints about mosaic mutations with low variant allele fraction is that they might be derived from circulating tumor cells (originated from cancer tissue) in blood of cancer patients. That is, the main point in the hot debate issues is that such mosaic variants might represent not bona fide variants, but somatic mutations originated by contamination from tumor tissues in cancer patients. In order to check whether our mosaic mutations might be originated from such contaminated circulating tumor cells in blood, we have compared our 23 mosaic mutations’ genomic locations with the non-TCGA data in the Exome Aggregation Consortium database, which do not include variation data from normal blood sample paired with TCGA clinical cancer sample from each patient.
We have identified that, of the 23 mosaic pathogenic mutations, 15 belonged to the non-TCGA data, corroborating that they are bona fide pathogenic mosaic mutations. Regarding the remaining 8 mosaic pathogenic mutations, whose genomic locations did not overlap with the non- TCGA data, we have confirmed that their genomic locations overlapped with the positions of previously known variants causing various hereditary disease syndromes. Also, the 15 pathogenic mosaic mutations overlapping with the non-TCGA data had been known to be involved in causing diverse hereditary disease syndromes This suggests that all of the 23 pathogenic mosaic mutations identified in this study are bona fide variants and that scientific communities should no longer ignore any pathogenic mosaic mutations with low VAFs as ones deserving no attention, from now on.
Keeping the above-mentioned critical viewpoint in mind, in this investigation we had explored mosaic pathogenic mutations using public germline variant database and the changes of their VAFs between normal blood and cancer tissues of breast cancer patients. Future detailed investigation of such VAF changes may provide us with a novel insight into how those mosaic pathogenic mutations, which might be potentially dangerous, but not phenotypically obvious before ill condition of their carriers, could facilitate predisposition to carcinogenesis through forming dominant tumorigenic expansion of somatic clones harboring those pathogenic mutations.
We surmise that the increasing changes in variant allele fraction of those pathogenic mutations between blood and cancerous tissues might be corresponding to the explanation for such tumorigenic clonal expansion.
So far, a majority of the mutations with the low VAFs (less than 30%, mainly) had been excluded in most of previous publications due to the ignorance of their importance. However, the exome aggregation consortium, which aimed to re-sequence blood samples and reannotate variation landscapes across diverse human clinical blood sample source types, for the first time, had openly published a largescale database of mutations with the low VAFs. Using those highquality mosaic mutation data in the exome aggregation consortium database, we have intersected them with the COSMIC database and TCGA BRCA data, and finally we have discovered 23 mosaic pathogenic mutations with low VAFs (less than 30%), which could play critical roles in causing predisposition to breast cancer, as well as diverse other cancer types and hereditary diseases. The fact that those mosaic pathogenic mutations occurred recurrently in diverse cancer types suggests that different cancer types might share some part of molecular mechanisms causing predisposition to carcinogenesis in their subtypes or as yet undefined subgroups.
Conclusion
From now on, if clinical cancer genomics community is to routinely and openly report mosaic mutations with low VAFs (less than 30%), the present shortage of the low VAF data for mosaic mutations will be overcome in the upcoming years. Furthermore, the abundant data of mosaic mutations with the low VAF that will be obtained in the upcoming years will contribute to elucidating novel predisposition mechanisms of carcinogenesis and clonal expansion caused by mutational mosaicism and also to revealing new diagnostic and therapeutic targets for detecting and treating the early stage of breast cancer, as well as diverse other cancer types.
Acknowledgments
This study has been supported by a research grant (2017R1D1A1B04033856 (Ryong Nam Kim)) funded by the Ministry of Education and the National Research Foundation in Korea.
References
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature 502: 333-339.
- Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, et al. (2012) Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med 4: 136-168.
- Friedman E, Efrat N, Soussan-Gutman L, Dvir A, Kaplan Y, et al. (2015) Low-level constitutional mosaicism of a de novo brca1 gene mutation. Br J Cancer 112: 765-768.
- Lavdovskaia ED, Iyevleva AG, Sokolenko AP, Mitiushkina NV, Preobrazhenskaya EV, et al. (2018) Egfr t790m mutation in tki-naive clinical samples: Frequency, tissue mosaicism, predictive value and awareness on artifacts. Oncol Res Treat 41: 634-642.
- Grzybowska E, Sieminska M, Zientek H, Kalinowska E, Michalska J, et al. (2002) Germline mutations in the brca1 gene predisposing to breast and ovarian cancers in upper silesia population. Acta Biochim Pol 49: 351-356.
- Hamann U, Liu X, Lange S, Ulmer HU, Benner A, et al. (2002) Contribution of brca2 germline mutations to hereditary breast/ovarian cancer in germany. J Med Genet 39: e12.
- Sung PL, Wen KC, Chen YJ, Chao TC, Tsai YF, et al. (2017) The frequency of cancer predisposition gene mutations in hereditary breast and ovarian cancer patients in taiwan: From brca1/2 to multi-gene panels. Plos One 12: e0185615.
- Wang YA, Jian JW, Hung CF, Peng HP, Yang CF, et al. (2018) Germline breast cancer susceptibility gene mutations and breast cancer outcomes. BMC Cancer 18: 315.
- Maxwell KN, Wubbenhorst B, Wenz BM, De Sloover D, Pluta J, et al. (2017) Brca locus-specific loss of heterozygosity in germline brca1 and brca2 carriers. Nat Commun 8: 319.
- Konishi H, Mohseni M, Tamaki A, Garay JP, Croessmann S, et al. (2011) Mutation of a single allele of the cancer susceptibility gene brca1 leads to genomic instability in human breast epithelial cells. Proc Natl Acad Sci USA 108: 17773-17778.
- Salmena L, Narod S (2012) Brca1 haploinsufficiency: Consequences for breast cancer. Womens Health (Lond) 8: 127-129.
- Marmorstein LY, Ouchi T, Aaronson SA (1998) The brca2 gene product functionally interacts with p53 and rad51. Proc Natl Acad Sci U S A 95: 13869-13874.
- Rajagopalan S, Andreeva A, Rutherford TJ, Fersht AR (2010) Mapping the physical and functional interactions between the tumor suppressors p53 and brca2. P Natl Acad Sci USA 107: 8587-8592.
- Ouchi T, Monteiro ANA, August A, Aaronson SA, Hanafusa H (1998) Brca1 regulates p53-dependent gene expression. P Natl Acad Sci U S A 95: 2302-2306.
- Ou YH, Chung PH, Sun TP, Shieh SY (2005) P53 c-terminal phosphorylation by chk1 and chk2 participates in the regulation of DNA-damage-induced c-terminal acetylation. Mol Biol Cell 16: 1684-1695.
- Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, et al. (1997) Role of phosphoinositide 3-oh kinase in cell transformation and control of the actin cytoskeleton by ras. Cell 89: 457-467.
- Meyer DS, Koren S, Leroy C, Brinkhaus H, Muller U, et al. (2013) Expression of pik3ca mutant e545k in the mammary gland induces heterogeneous tumors but is less potent than mutant h1047r. Oncogenesis 2: e74.
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, et al. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123: 725-731.
- Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, et al. (2018) Pathogenic germline variants in 10,389 adult cancers. Cell 173: 355-370 e314.
- Rajendran BK, Deng CX (2017) Characterization of potential driver mutations involved in human breast cancer by computational approaches. Oncotarget 8: 50252-50272.
 Spanish
Spanish  Chinese
Chinese  Russian
Russian  German
German  French
French  Japanese
Japanese  Portuguese
Portuguese  Hindi
Hindi